Figures & data
Figure 1. Calculated cumulative potential energy (U) for Aβ42 in aqueous solution with a water layer of 20 Å (dashed line) and 30 Å (dotted line) for 160 ns, respectively (A); correlation of calculated and experimental Cα chemical shift values obtained from Dr. Michael Zagorski for Aβ42 in an aqueous solution with a water layer of 20 Å (B) and 30 Å (C), respectively.
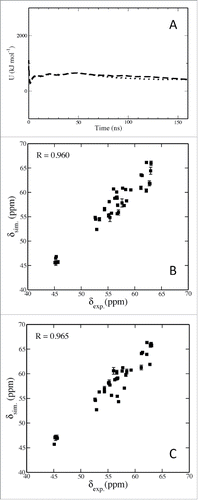
Table 1. Protocols and standards used in our different simulations.
Figure 2. Enthalpic (A) and entropic (B) contributions to the Gibbs free energy values of monomeric Aβ42 conformations in aqueous solution with Vs = 330 nm3 (blue) and Vs = 810 nm3 (red) where Vs presents the volume of water in the solution.
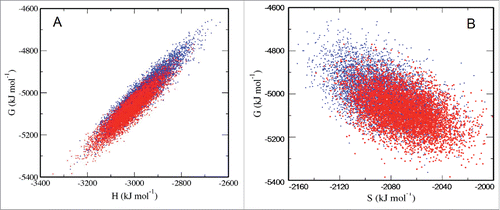
Figure 3. Differences in secondary structure abundances in monomeric Aβ42 in aqueous solution with an increase in confined aqueous volume, corresponding to an increase from Vs = 330 nm3 to Vs = 810 nm3 (Vs is the volume of water).
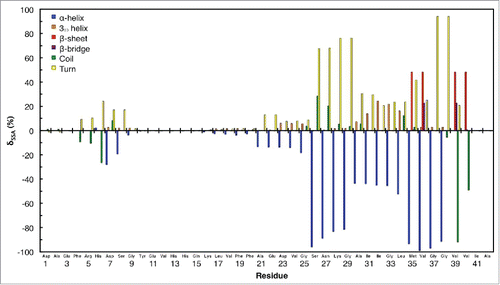
Figure 4. The intra-molecular peptide interaction (I), intra-molecular hydrogen bond (II), and hydrophobic interactions (III) maps for the Aβ42 monomer in aqueous solution with Vs = 330 nm3 (A) and Vs = 810 nm3 (B) where Vs presents the volume of water.
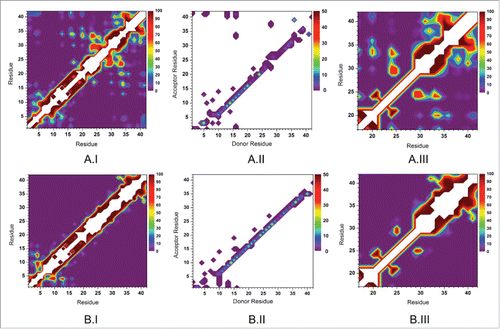
Table 2. Formed salt bridges along with their abundances in monomeric Aβ42 in aqueous solution with different confined aqueous volume effects, corresponding to water volumes of 330 nm3 and 810 nm3, respectively.
Figure 5. Convergence of Aβ42 simulations via REMD simulations tested using the secondary structure abundances with time.
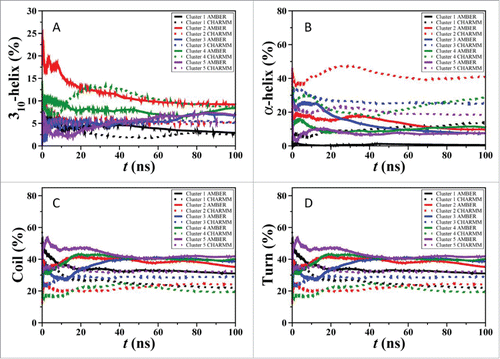
Figure 6. Radius of gyration values using AMBER FF99SB and CHARMM22/CMAP parameters in an implicit model of water. (A) Cluster 1, (B) Cluster 2, (C) Cluster 3, (D) Cluster 4, (E) Cluster 5.
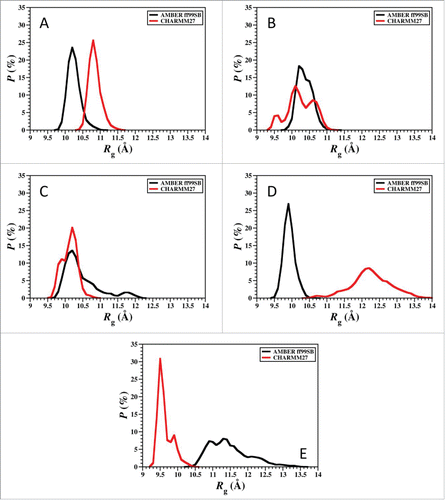
Figure 7. Secondary structure properties simulated using AMBER FF99SB (Black) and CHARMM22/CMAP (Red) parameters in an implicit water environment; (A) Cluster 1, (B) Cluster 2, (C) Cluster 3, (D) Cluster 4, (E) Cluster 5.
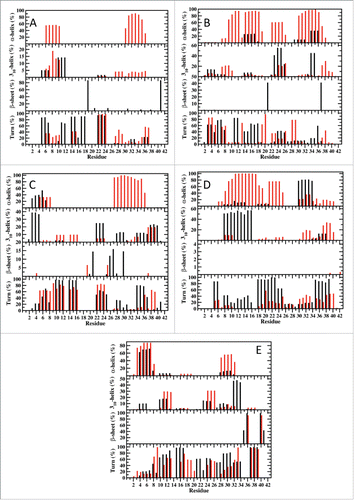
Figure 8. Tertiary structure properties simulated using AMBER FF99SB and CHARMM22/CMAP parameters in an implicit water environment. (A) Cluster 1 using AMBER FF99SB parameters, (B) Cluster 1 using CHARMM22/CMAP parameters, (C) Cluster 2 using AMBER FF99SB parameters, (D) Cluster 2 using CHARMM22/CMAP parameters, (E) Cluster 3 using AMBER FF99SB parameters, (F) Cluster 3 using CHARMM22/CMAP parameters, (G) Cluster 4 using AMBER FF99SB parameters, (H) Cluster 4 using CHARMM22/CMAP parameters, (I) Cluster 5 using AMBER FF99SB parameters, (J) Cluster 5 using CHARMM22/CMAP parameters.
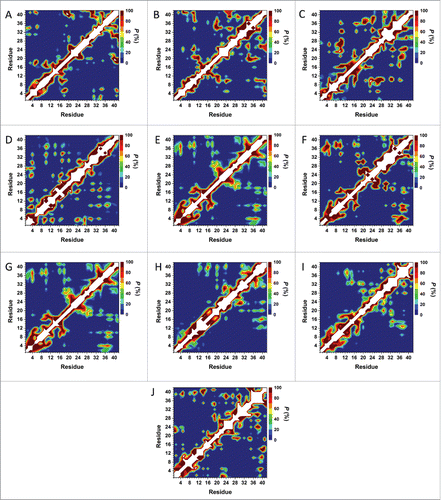
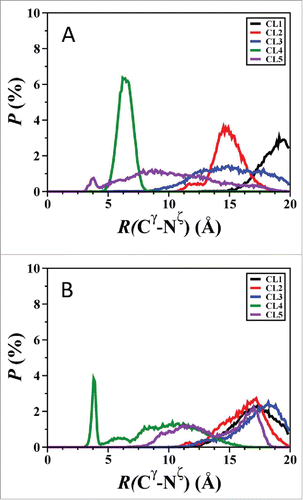
Figure 9. The calculated probability distribution of the distance between the Cγ atom of the Asp23 residue and the Nζ atom of the Lys28 residues for the Aβ42 using AMBER FF99SB and CHARMM22/CMAP parameters in an implicit water environment. Simulations were conducted using AMBER FF99SB (A) and CHARMM22/CMAP parameters (B).