Figures & data
Figure 1. 2-oxoglutarate-oxygenase catalysis.(A) Catalytic cycle. Catalysis requires essential co-factors Fe(II), molecular oxygen (O2), and the Krebs cycle intermediate 2-oxoglutarate (2OG), together with the ‘2-His 1-carboxylate’ motif (His-Asp-His) within the active site of the enzyme. Note that one atom of oxygen from molecular oxygen is incorporated into the product, and that the reaction generates succinate and carbon dioxide. (B) 2OG-oxygenases catalyze stable hydroxylation (blue box) and demethylation via hydroxylation (green box) of DNA, RNA, lipid and protein. Note that ascorbate is required for full activity of a subset of 2OG-oxygenases (hence the smaller font). Hydroxylation of a methyl group generally creates a highly unstable hydroxymethyl intermediate that decomposes into formaldehyde (CHOH) and the unmodified residue. The fate of the formaldehyde is not known, but may be metabolized by formaldehyde dehydrogenase. Note that in some chemical contexts hydroxylation of a methyl group can create a stable hydroxymethyl product, such as that catalyzed by TETs. The oncometabolite 2-hydroxyglutarate (2HG) can interfere with 2OG-oxygenase function by acting as an activating co-substrate in some instances, or as a 2OG competitive inhibitor in others. Succinate and fumarate inhibit 2OG-oxygenases by product inhibition and 2OG competition, respectively.
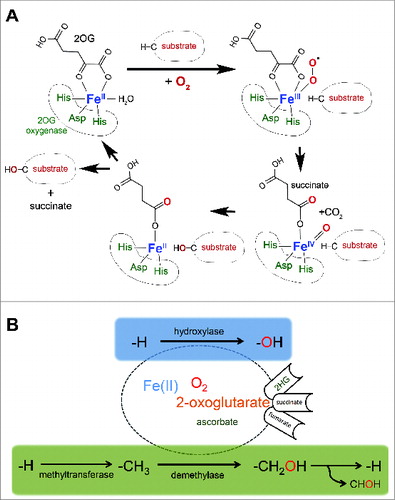
Table 1. 2OG-oxygenases with targets in ribosome biology and protein synthesis are frequently implicated in disease, particularly cancer. It should be noted that other substrates of these enzymes may exist in other biological contexts and that the critical targets of these enzymes involved in disease are often unclear, but may include the translational targets listed. The role of JmjC family 2OG-oxygenases in disease was recently reviewed by Oppermann and colleaguesCitation10
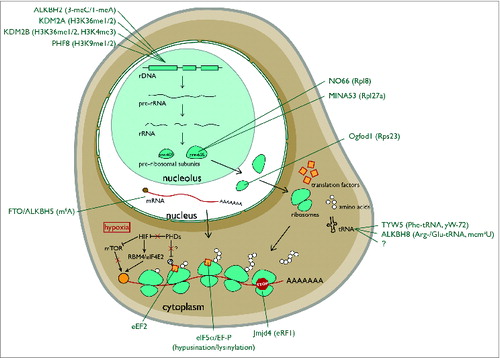
Figure 2. Modifications catalyzed by 2OG-oxygenases in ribosome biology. 2OG-oxygenases are in bold above the arrows, with the corresponding substrate in brackets underneath. Upper panel: Modifications to protein. The modifications presented in the top row represent histone lysine demethylation by various JmjC histone demethylases. H3K=Histone H3 lysine. me=methyl group. The second and third rows in this panel represent stable hydroxylation of amino acid side chains by the indicated 2OG-oxygenases. Lower panel: Modifications to RNA. Presented in the same format as the upper panel. 5mC=5-methylcytosine. 5hmC=5-hydroxymethylcytosine. mcm5U=methoxycarbonylmethyluridine. mchm5U=methoxycarbonylhydroxymethyluridine. yW=wybutosine. yW-72=wybutosine minus 72Da. OHyW*=undermodified hydroxywybutosine. OHyW=hydroxywybutosine, formed by attachment of methyl and methoxycarbonyl groups to the aminocarboxyl side chain of OHyW* by TYW4.
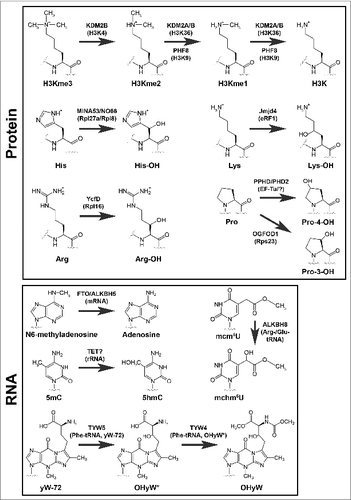
Figure 3. Hydroxylation and demethylation events in eukaryotic ribosome biogenesis and protein translation. ALKBH2 is a demethylase that repairs alkylated rDNA. 3-meC=3-methylcytosine. 1-meA=1methyladenine. KDM2A/B and PHF8 are nucleolar histone lysine demethylases that target rDNA. MINA53 and NO66 are nucleolar histidyl hydroxylases of the large ribosomal subunit. Ogfod1 is a nuclear prolyl hydroxylase of the small ribosomal subunit. FTO and ALKBH5 are m6A RNA demethylases. TYW5 and ALKBH8 hydroxylate the anti-codon loop of the indicated tRNAs. The ‘?’ under ALKBH8 denotes an as yet unidentified mcm5U hydroxylase. Jmjd4 is a hydroxylase of the translational termination factor eRF1. Note that hypoxia (red box) substantially regulates translation. Inhibition of prolyl hydroxylases in hypoxia indirectly represses EIF4E (via HIF-dependent mTOR inhibition) while activating the translation of specific transcripts via an RBM4/HIF2α/eIF4E2 cap-dependent mechanism. The orange ball represents the cap.