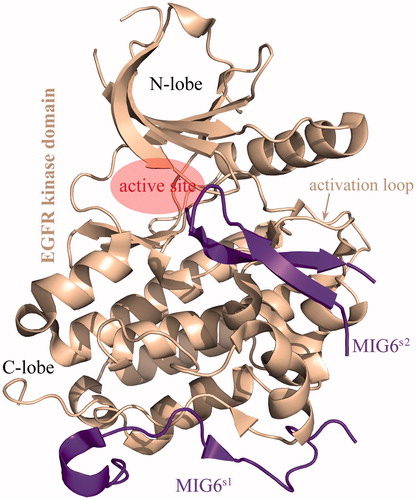
Figures & data
Figure 2. (A) The phosphorylated MIG6s2 peptide was stripped from its cocrystallizated complex structure with EGFR kinase domain (PDB: 4ZJV), resulting in unbound phosphorylated peptide with the conformation as it in cocrystallizated complex. (B) The stripped peptide was dephosphorylated at residues pTyr394 and pTyr395 without conformation change, resulting in a dephosphorylated version of the peptide. (C) The Leu397 residue of the stripped peptide was virtually mutated to Cys397, which was then covalently connected to wild-type Cys378 residue by computationally modeling a disulfide bond between them, resulting in a phosphorylated cyclic peptide. Subsequently, the three artificially modeled peptides were subjected to MD simulations to reach at equilibrium state.
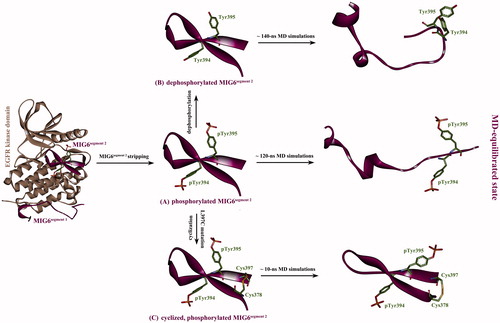
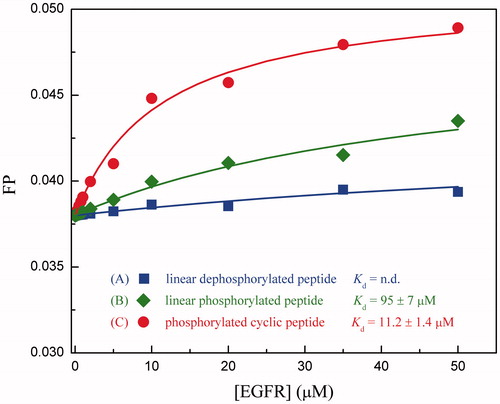
Table 1. The calculated energetics parameters and experimental affinities for the different forms of MIG6s2 peptides to interact with EGFR kinase domain.
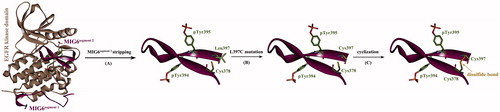
Figure 3. Cyclization of phosphorylated MIG6s2 peptide. (A) The phosphorylated peptide was stripped from its cocrystallizated complex with EGFR kinase domain (PDB: 4ZJV). (B) The Leu397 residue of the peptide was virtually mutated to Cys397 (L397C). (C) The peptide was cyclized by introducing a disulfide bond between the mutated Cys397 and wild-type Cys378.