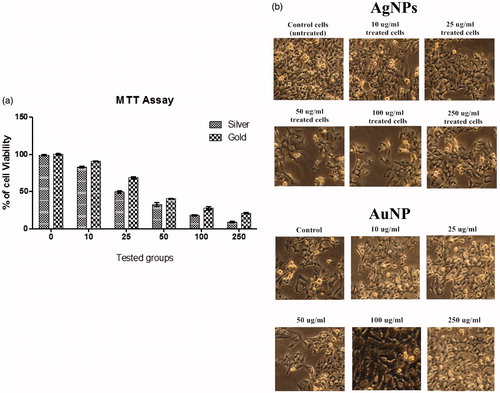
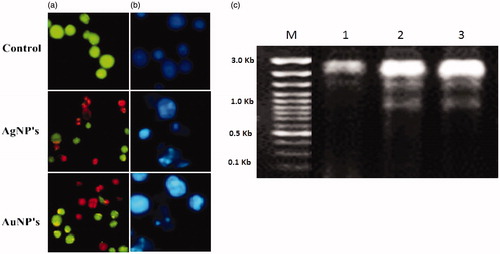
Figures & data
Figure 1. Neighbor-joining phylogenetic tree based on 16S rRNA gene sequence analysis using the maximum parsimony method in MEGA ver. 6.0. showing phylogenetic relationships of strain X. stockae KT835471 and members of the genus Xenorhabdus.
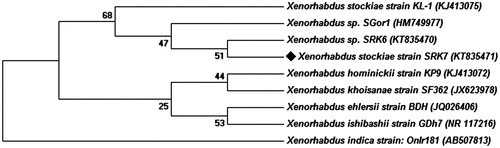
Figure 2. (a) Synthesis of Ag and AuNPs. (1). Culture supernatants of X. stockae KT835471 (2). AgNPs and (3). AuNPs and (b) UV–spectroscopic analysis of synthesized AgNPs and (c) AuNPs shows the SPR peak at 430 nm and 560 nm, respectively.
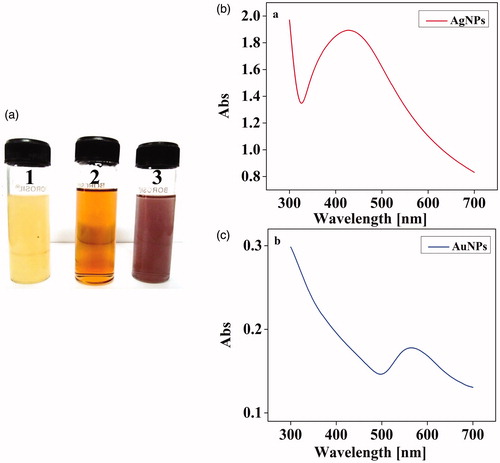
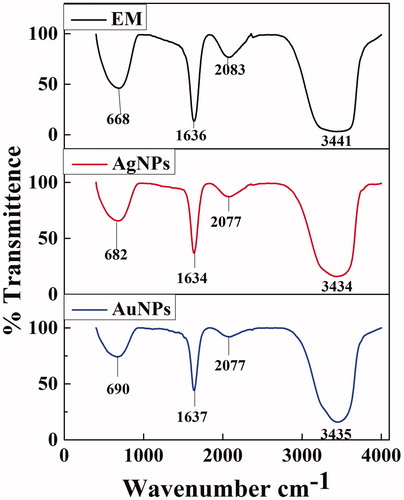
Figure 4. X-ray Diffraction analysis of synthesized (a) XsAgNPs and (b) XsAuNPs shows the corresponding diffraction value of face centered cubic crystalline structures.
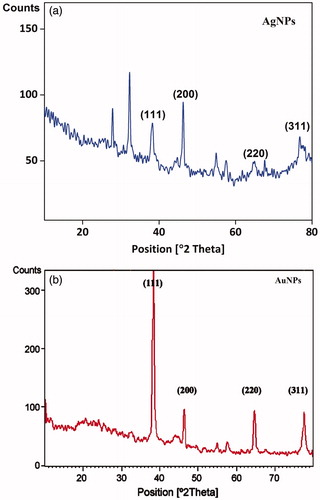
Figure 5. (a) HRTEM micrographs displays well-distributed spherical AgNPs with an average size of 14 ± 6 nm (b) EDAX analysis gives a strong signal for Ag and (c) Particle size distribution of XsAgNPs.
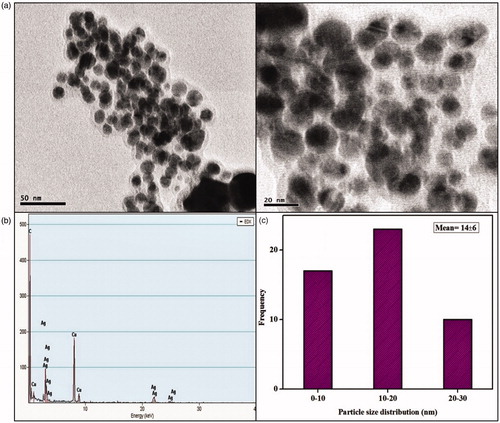
Figure 6. (a) HRTEM micrographs displays well-distributed spherical and triangle AuNPs with an average size of 14 ± 5 nm (b) EDAX analysis gives a strong signal for Au and (c) Particle size distribution of XsAuNPs.
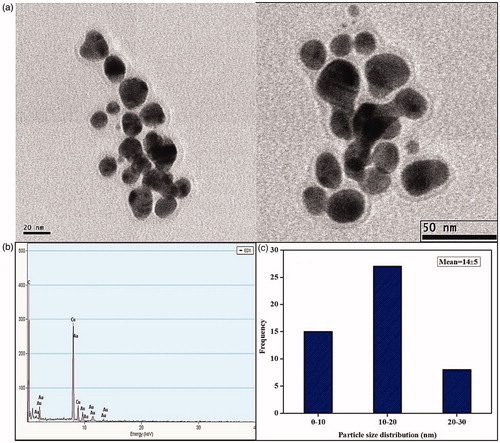
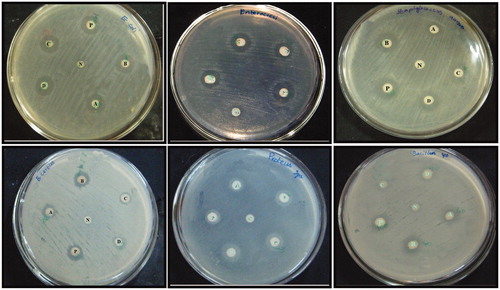
Figure 7. A clear zone of inhibition indicates the bactericidal action of synthesized NPs (A) 0.20 μl of AgNPs; (B) 40 μl of AgNPs; (C) 0.20 μl of AuNPs; (D) 40 μl of AuNPs; P-Positive control; N-Negative control.