Figures & data
Scheme 1. Synthetic pathway of 5-(4-trifluoromethylphenylsulphonamido)benzoxazole derivatives. Reagents and conditions: (A) PPA, 170–200 °C, 1.5–2.5 h; (B) Pyridine and dichloromethane, RT, 16 h.
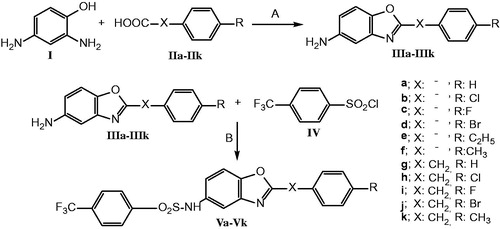
Figure 1. (a) Binding sites of dimeric hGST P1-1 enzyme (pdb ID 2GSS and 6GSS): the protein structure is represented in solid ribbon secondary type. (b) Docking pose of compound 5f and Vh in 6GSS. (c) Docking pose of Vh*: sulphonyl group revealed H bonds with SH and glycine NH of GSH, CF3 group of the compound showed halogen bond with Cys101, compound had interactions with Pro9 (alkyl hydrophobic), Val35 (pi-alkyl), Ile104 (pi-alkyl), and Tyr108 (pi–pi). (d) Docking pose of 5f*: sulphonyl group revealed H bond with Gln51 and Tyr108, compound had interactions with Arg13 (electrostatic), Lys44 (alkyl hydrophobic), Tyr108 (pi–pi), and GSH (pi-sulfur). (e) Docking pose of Vd*: sulphonyl group revealed H bonds with Tyr108 and glycine NH of GSH and CF3 group revealed H bond with Tyr109, compound had interactions with Phe8 (pi–pi), Val10 (pi-alkyl), Val35 (pi-alkyl), and Tyr108 (pi–pi). (f) Docking pose of Vk*: sulphonyl group revealed H bond with Glycine NH of GSH, CF3 group of the compound showed halogen bond with Ile104, compound had interactions with Phe8 (pi–pi), pro9 (pi-alkyl and alkyl hydrophobic), Ile104 (pi-alkyl and alkyl hydrophobic), Tyr108 (pi–pi), and GSH (pi-sigma). *The protein structure is represented in flat ribbon style, GSH and all the ligands are seen in stick representation.
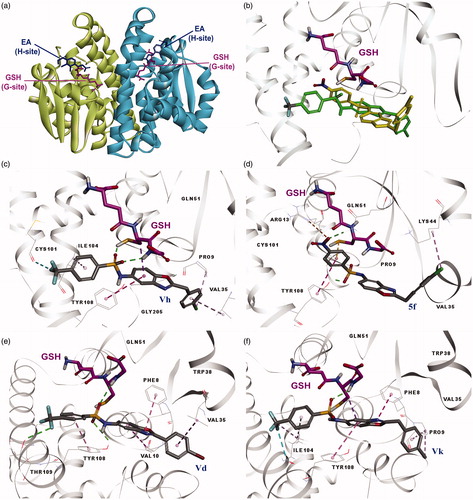
Table 1. Docking results.
Table 2. The structures of the tested sulfonamidobenzoxazoles and their in vitro hGST P1-1 inhibitory effects.
