Figures & data
Figure 1. The overall graphical representation of the Anemarrhena asphodeloides mediated eco-friendly synthesis of silver (Aa-AgNPs) and gold (Aa-AuNPs) nanoparticles.
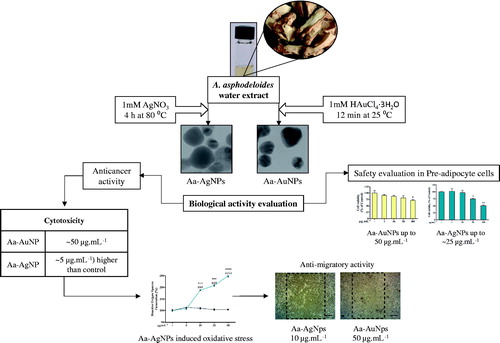
Figure 2. UV-visual spectrophotometry indicated the synthesis of Aa-AuNPs and Aa-AgNPs. For the synthesis of Aa-AgNPs incubation for 4 h (a) at 80 °C (b) gave the best results, while for Aa-AuNPs synthesis the optimal conditions were at for 12 min (c) incubation at room temperature (d).
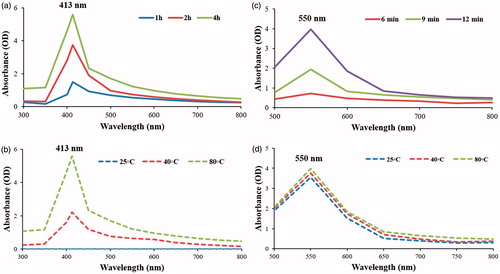
Figure 3. FE-TEM uncovered that both Aa-AgNPs (i(a,c)) and Aa-AuNPs (ii(a,c)) are predominantly spherical. SAED pattern indicated the crystallinity of both Aa-AgNPs (i(b)) and Aa-AuNPs (ii(b)). Furthermore, the purity of Aa-AgNPs (i(d,e)), Aa-AuNPs (ii(d,e)) were determined by elemental mapping and EDX analyses. Scale bar: (i(a)) and (ii(a)) – 5 nm; (i(c)) and (i(d)) – 80 nm; (ii(c)) and (ii(d)) – 70 nm.
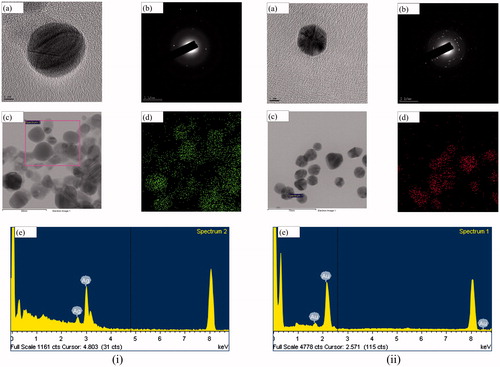
Figure 4. DLS analysis indicated the particle size distribution based on intensity of both Aa-AgNPs (a) and Aa-AuNPs (b).
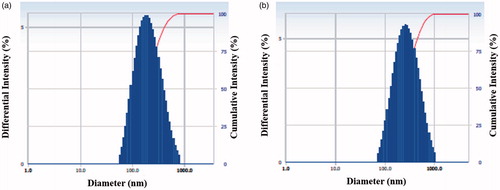
Figure 5. The crystallinity of both Aa-AgNPs (a) and Aa-AuNPs (b) were confirmed by XRD analysis. In addition, the biological molecules attached to Aa-AgNPs and Aa-AuNPs were determined by FTIR (c). The FTIR pattern of the extract is also indicated.
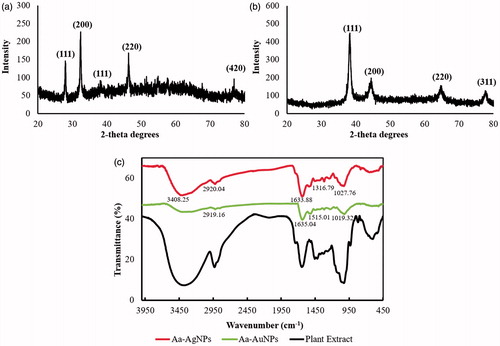
Figure 6. Cytotoxicity of Gold (Aa-AuNPs). Cell viability of (a) 3T3-L1, Pre-adipocyte cells; (b) A549, lung cancer cells; (c) HT29, colon cancer cells; (d) MCF7, breast cancer cells was measured after 48 h of treatment by MTT assay. Data are shown as mean ± SEM (n = 3). *p < .05, **p <.01 and ***p < .001 vs. Control.
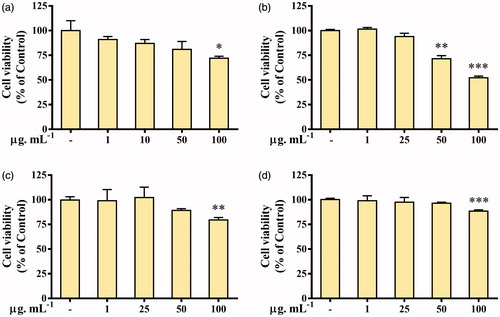
Figure 7. Cytotoxicity of green Silver (Aa-AgNPs) Nanoparticles and commercial Silver (cAgNPs) Nanoparticles. Cell viability was measured after 48 h of treatment by MTT assay in (a) 3T3-L1, Pre-adipocyte cells; (b) A549, lung cancer cells; (c) HT29, colon cancer cells; (d) MCF7, breast cancer cells. Data are shown as mean ± SEM (n = 3). *p < .05, **p < .01 and ***p < .001 vs. Control. ###p < .001 Aa-AgNPs vs. cAgNPs.
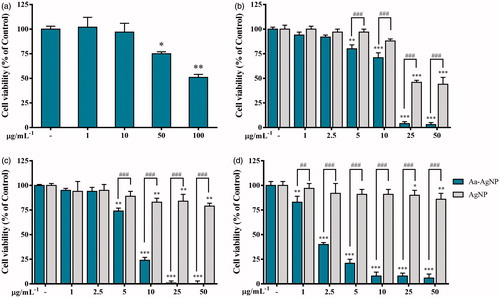
Figure 8. Oxidative stress potential of green Silver (Aa-AgNPs) Nanoparticles and commercial Silver (cAgNPs) Nanoparticles. Reactive oxygen species generation was measured after 48 h of treatment in (a) A549, (b) HT29 and (c) MCF7 cells. Data are shown as mean ± SEM (n = 3). *p < .05, ***p < .001 vs. Control. ###p < .001 Aa-AgNPs vs. cAgNPs.
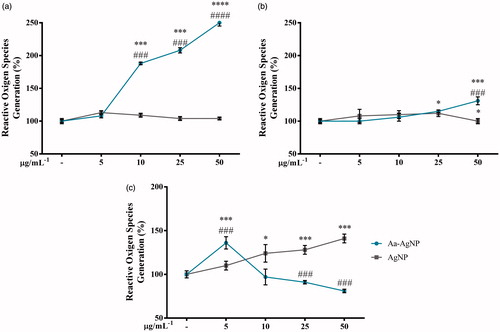
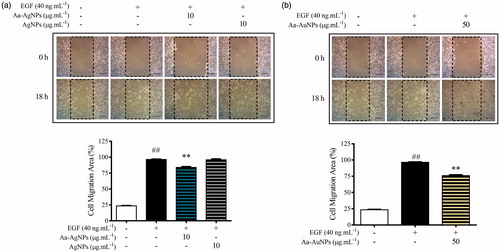
Figure 9. Aa-AgNPs reduces EGF-enhanced migration of A549 lung cancer cells. (a) Gold (Aa-AuNPs) Nanoparticles and (b) Silver (Aa-AgNPs) Nanoparticles analysis of migration ratio (%) within 24 h in wound healing assay. Data are shown as mean ± SEM (n = 3). ###p < .01 vs. Control. **p < .01 vs. EGF-Control.