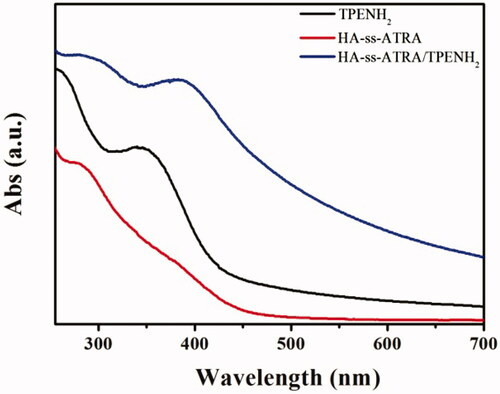
Figures & data
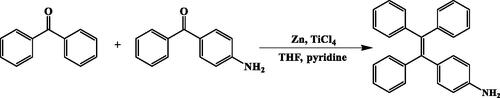
Scheme 1. Synthesis of HA-ss-ATRA/TPENH2 conjugates (A). Illustration of the self-assembly, accumulation at cancer cells, intracellular trafficking pathway of redox-sensitive AIE HNPs, and real-time intracellular imaging of AIE HNPs (B). Circulation of AIE HNPs within vessel (a); Accumulation of DOX-loaded HNPs within the tumor site through passive and active targeting effects (b); CD44 receptor-mediated cellular internalization (c, d); Endo/Lysosomal escape, and GSH-controllable release of DOX in cytoplasm (e, f); Released DOX into nucleus for apoptosis and cytotoxicity (g).
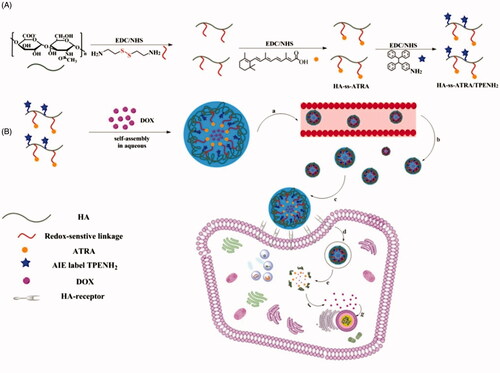
Figure 3. Representative SEM images (A) and TEM images (B) of HA-ss-ATRA/TPENH2 HNPs. size distribution of HA-ss-ATRA/TPENH2 HNPs (C).
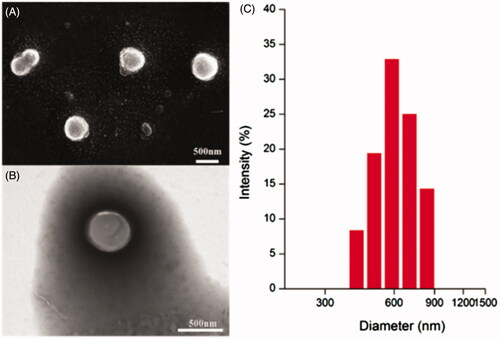
Figure 4. GSH triggered DOX release from HA-ss-ATRA/TPENH2 HNPs and HA-ATRA/TPENH2 HNPs. The error bars in the graph represent standard deviations (n = 3).
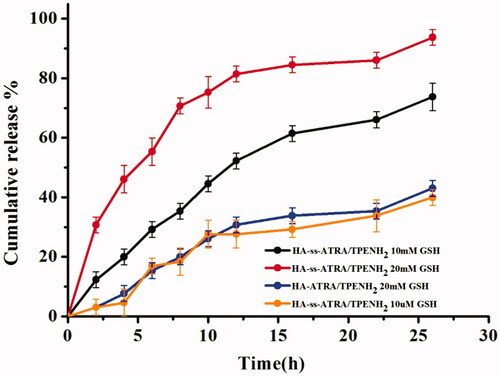
Figure 5. Cell viability of HepG-2 cells treated with DOX-loaded HA-ss-ATRA/TPENH2 HNPs and free DOX after 24 h (A). Cell viability of HepG-2 cells treated with DOX-loaded HA-ss-ATRA/TPENH2 HNPs and free DOX after 24 h, 48 h and 72 h (B). Cytotoxicity of blank HA-ss-ATRA/TPENH2 HNPs against HepG-2 cells and 293T cells (C). Cell viability of HepG-2 cells treated with DOX-loaded HA-ss-ATRA/TPENH2 HNPs and DOX-loaded HA-ATRA/TPENH2 HNPs (D). Cell survival fractions were assessed by MTT assay. Data represent mean ± SD (n = 5).
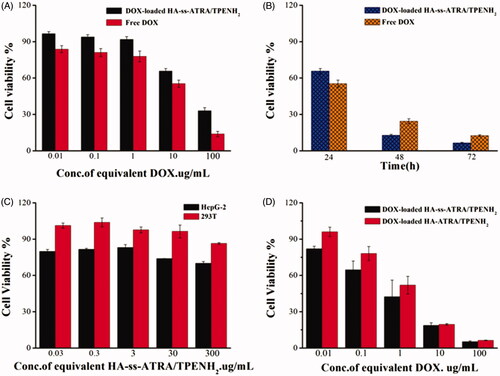
Table 1. IC50 of Free DOX and DOX-loaded HNPs for HepG-2 cells.
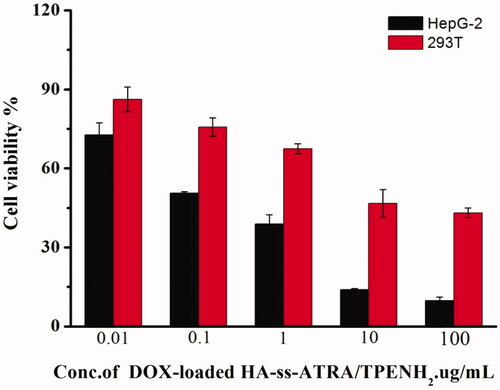
Figure 6. Cell viability of HepG-2 and 293T cells treated with DOX-loaded HA-ss-ATRA/TPENH2 HNPs after 48 h.