Figures & data
Figure 1. Structure of coronavirus spike glycoprotein and spike glycoprotein-ACE2 receptor complex spike glycoprotein mediates SARS-CoV-2 entry into the host cells. SARS-CoV-2 spike protein binds to its receptor ACE2 through its receptor-binding domain (RBD). The figure shows the closed and open conformations of the spike glycoprotein structure and spike protein–ACE2 receptor complex, respectively. The RBD (receptor binding domain) region binds to the ACE2 receptor, allowing the closed conformation to transition into an open conformation (created with BioRender.com).
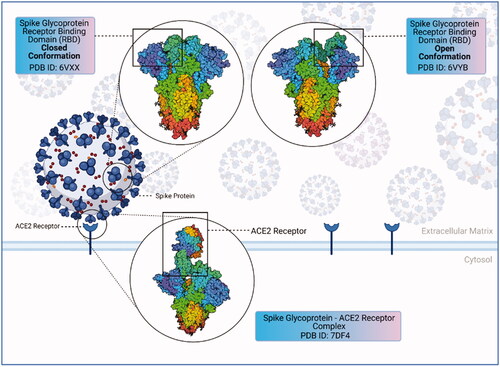
Figure 3. Secondary structure of a trans-cleaving hammerhead ribozyme with cleavage site (created with BioRender.com).
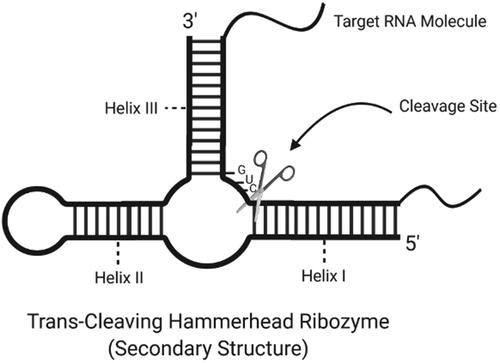
Figure 4. Secondary structure of a trans-cleaving HDV ribozyme with an estimated cleavage site and helix regions (created with BioRender.com).
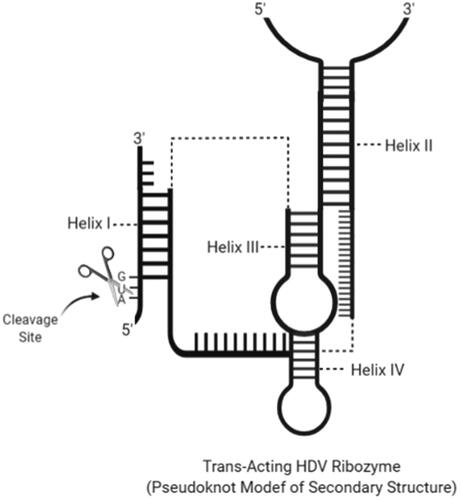
Figure 5. Secondary structure of a trans-cleavage hairpin ribozyme with cleavage site and helix regions (created with BioRender.com).
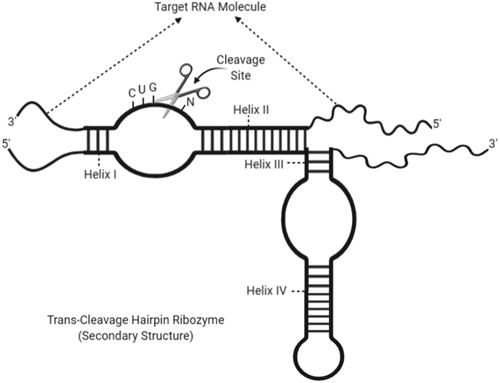
Figure 6. The secondary structure of a twister ribozyme with cleavage site and twisted regions (created with BioRender.com).
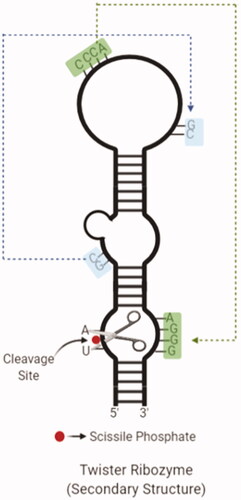
Figure 7. The secondary structure of an NVS ribozyme with cleavage site and helix regions (created with BioRender.com).
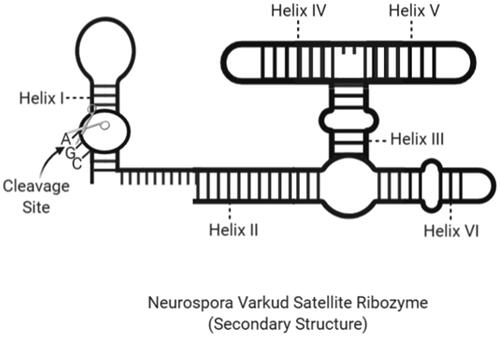
Figure 8. The secondary structure of a trans-cleavage GlmS ribozyme with cleavage site and helix regions (created with BioRender.com).
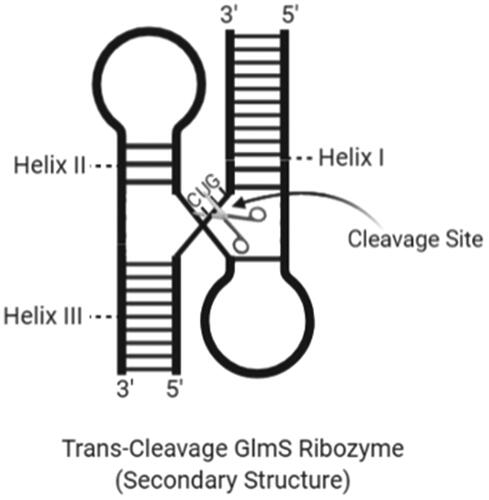
Figure 9. Light blue boxes indicate the open reading frame (ORF1a/b) of 5′UTR encoding the non-structural proteins required for viral replication. S box: spike protein; E box: envelope protein; M box: membrane protein; N box: nucleocapsid protein; yellow boxes: other ORFs.
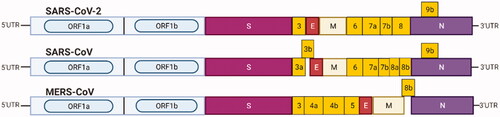
Figure 10. “Endogenous Expression": transcription of ribozyme-expressing vectors and synthesis of ribozymes (created with BioRender.com).
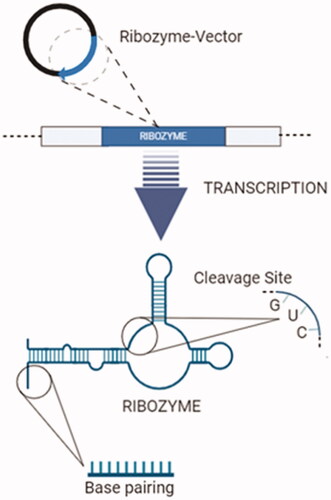
Figure 11. Transfer of ribozymes to the target cell via nanoparticle, a non-viral transfection tool. (1) Plasmid vectors that will express ribozyme. (2) Pre-synthesized ribozyme. (3) Synthesis of nanoparticles targeted to the ACE-2 receptor. (4) Conjugates formed by nanoparticles with ribozyme-vector and ribozymes. Conjugates are ready for target cell-specific transfection (created with BioRender.com).
Table 1. Clinical trials and results using anti-HIV-1 OZ1 ribozymes.
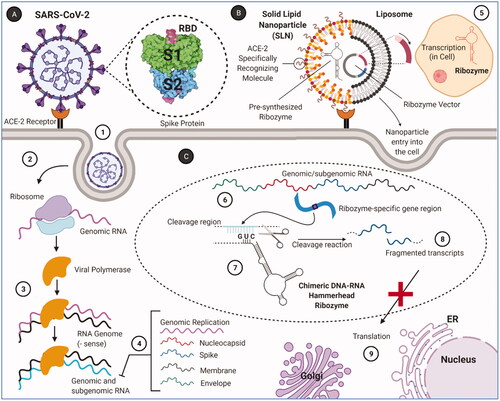
Figure 12. RNA replication of SARS-CoV-2 and target cleavage mechanism of target-specific ribozymes. (A) SARS-CoV-2 and intracellular replication process. (1) SARS-CoV-2 enters the cell through the endocytic pathway by binding to the ACE2 receptor. (2) The viral polymerase is translated by the ribosomes, following by the viral genome released. (3) Viral RNA is replicated by viral polymerase. (4) Genomic and subgenomic (nucleocapsid, spike, membrane, envelope) RNAs are produced. (B) Binding and entry of targeted ribozyme–nanoparticle conjugates to the target cell. (5) Ribozyme vectors that form the ribozyme–nanoparticle conjugate replicate inside the cell. (C) Cleavage reaction mechanism of ribozymes. (6) Genomic/subgenomic RNAs replicating in the cell contain specific regions to ribozyme binding. (7) These ribozyme-specific regions are cleaved after specific pairing. (8) Target RNAs are fragmented with the cleavage reaction. (9) Viral proteins are not formed due to fragmented transcripts cannot be translated (created with BioRender.com).