Figures & data
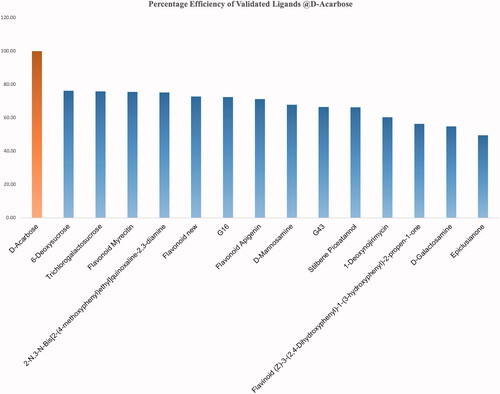
Table 1. Details of the validated ligands: the compound name refers to the compound used in the respective study.
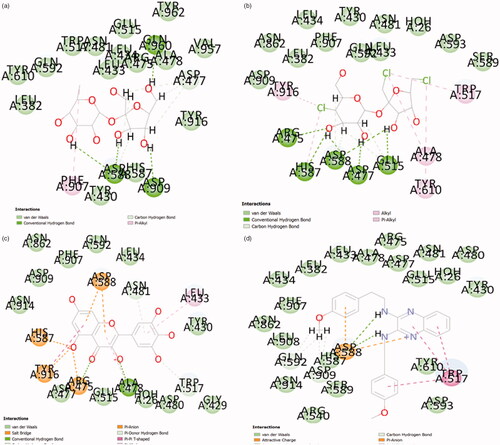
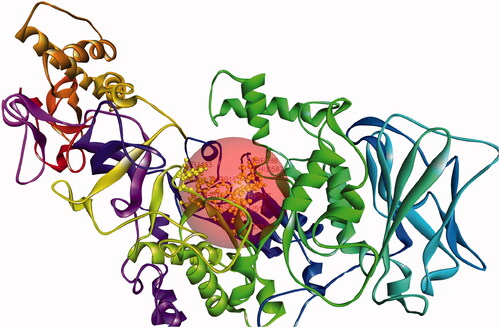
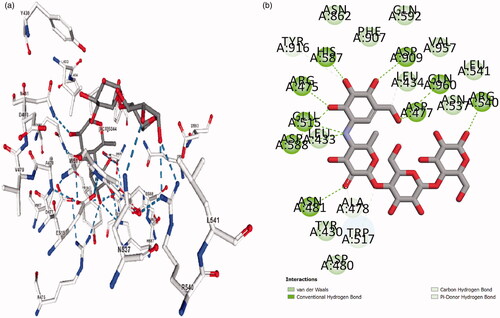
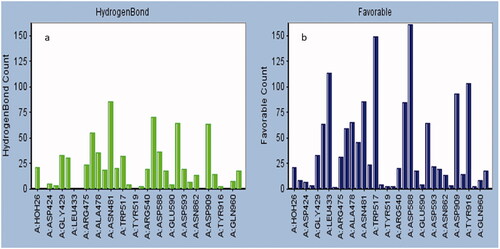
Table 2. Summary of ligand interactions.
Table 3. Summary of Interacting chemical fragments.
![Figure 1. GtfC reaction scheme (Brenda enzyme database) [Citation11].](/cms/asset/a158be20-b9a0-43bd-b4de-f4db798870ad/ianb_a_1903021_f0001_c.jpg)