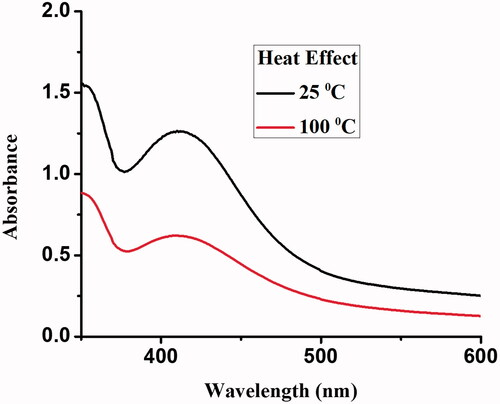
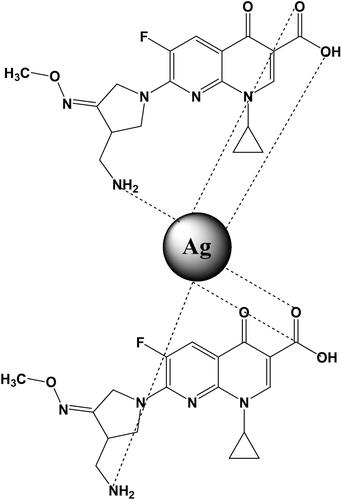
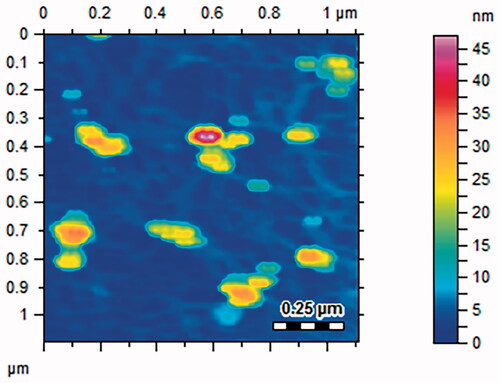
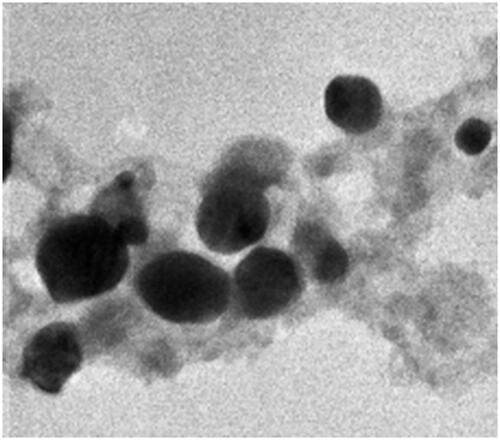
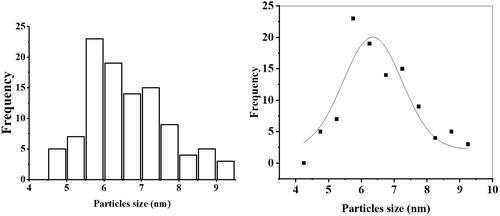
Figures & data
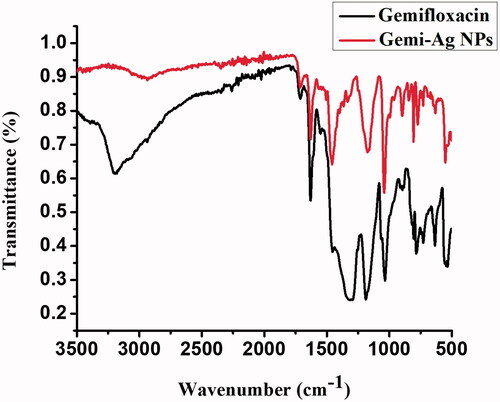
Table 1. FTIR spectral assignment of free gemifloxacin and Gemi-AgNPs.
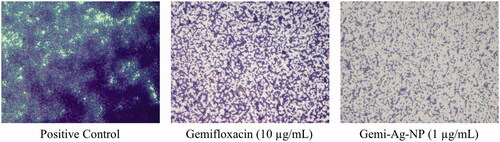
Table 2. Biofilm inhibition of gemifloxacin and Gemi-AgNPs.
Table 3. Biofilm eradication of gemifloxacin and Gemi-AgNPs.
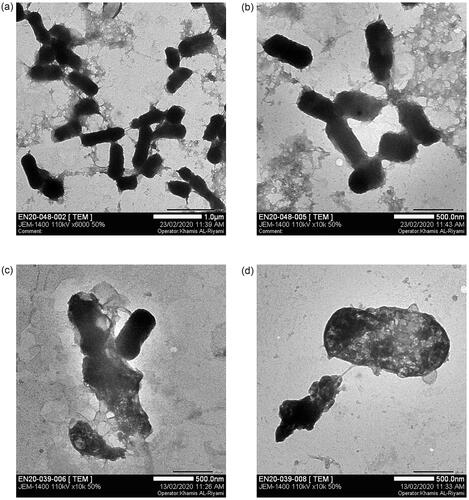
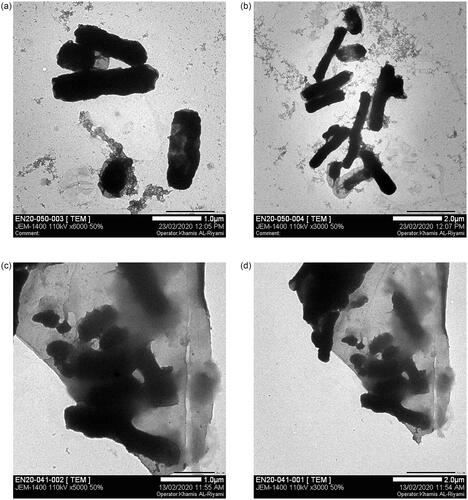
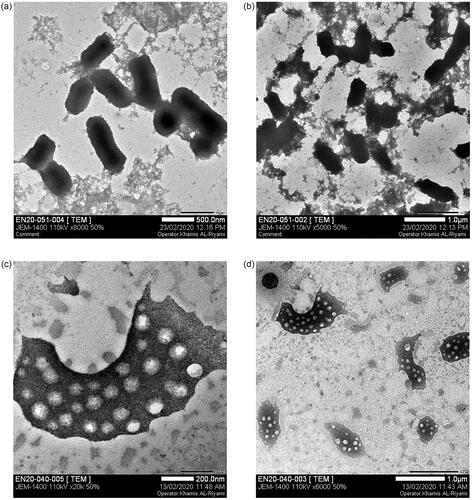
Figure 12. Cell wall damage of K. pneumoniae. (a) Untreated K. pneumoniae at 500 nm and (b) at 1 µm was used as control, whereas (c) present the bacterial cell wall damage of K. pneumoniae after treatment with Gemi-AgNPs at 200 nm and (d) at 1 µm.
Data availability statement
All data is included in the submission/manuscript file.