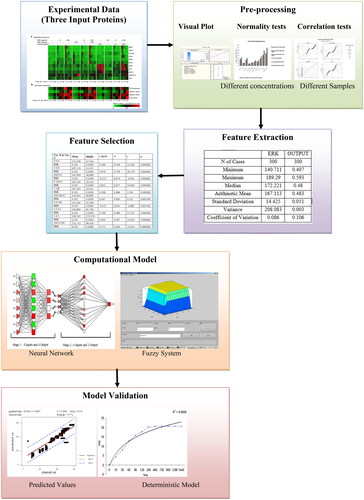
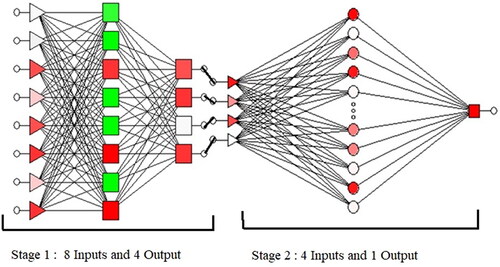
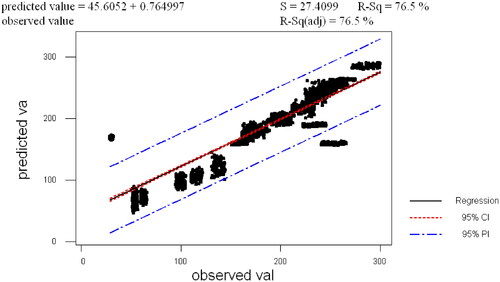
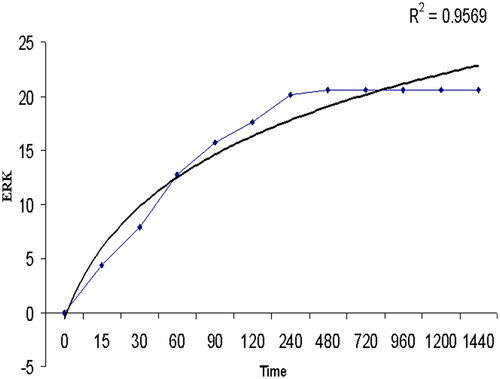
Figures & data
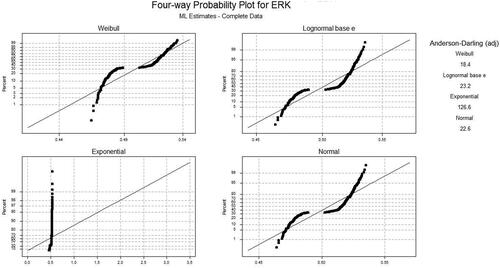
Figure 2. Statistical summary of Extracellular Signal Regulated Kinase protein (a) Kolmogorov-Smirnov plot (b) P-P plot, (c) various statistics values and (d) box plot.
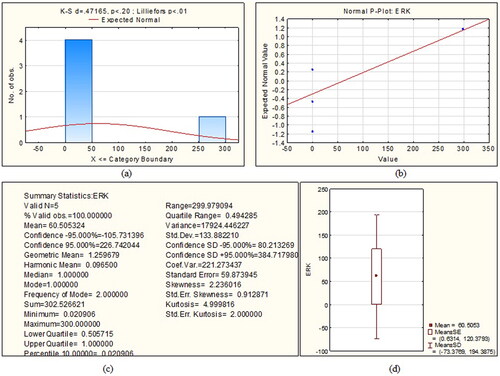
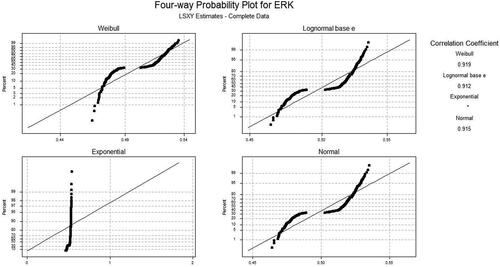
Figure 3. The normal, Kaplan Meier (KM) and Herd Johnson (HJ) method values for the Least Square (LS), and Maximum Likelihood (ML) technique.
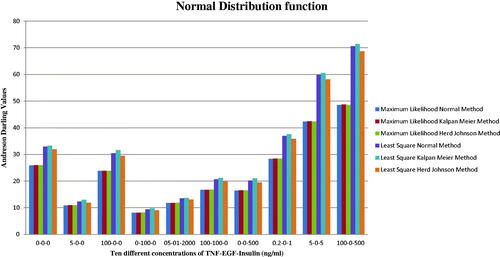
Table 1. Different concentrations of tumour necrosis factor (TNF)-epidermal growth factor (EGF)-insulin.
Table 2. Algorithm for the detection of cell survival/ death.
Table 3. Pearson correlation coefficient (PCC) for least square method.
Table 4. Correlation values for extracellular signal regulated kinase protein.
Table 5. Kaplan Meier, Normal, and Herd Johnson values using maximum likelihood of the different distribution functions for Extracellular Signal Regulated kinase protein.
Table 6. Anderson Darling values for different distribution functions based on different samples.
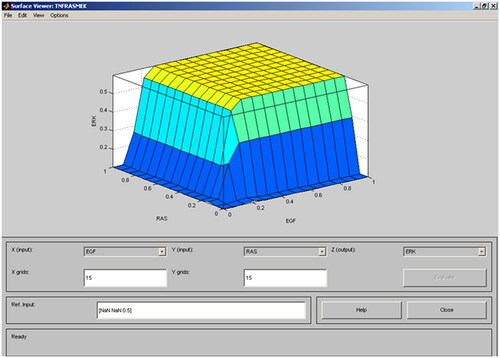
Table 7. Comparison of Anderson Darling values for the two different computational models.
Table 8. Different features of Extracellular Signal Regulated Kinase (ERK).
Data availability statement
The data analysed during the current study are not publicly available but is available from the corresponding author on reasonable request.