Figures & data
Table 1. Demographic and clinical characteristics of 60 renal transplant recipients infected by C. neoformans/C. gattii species complexes.
Figure 1. Molecular type distribution of 82 clinical isolates of C. neoformans/C. gattii species complexes (A) cultured from 60 renal transplants recipients (B).
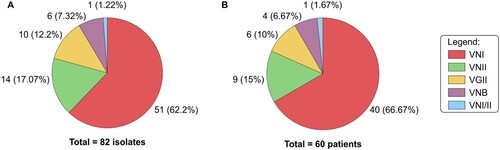
Figure 2. Phylogenetic relationships as inferred from a maximum likelihood analysis of CAP59, LAC1, PLB1, SOD1, URA5, TEF1 and IGS1 sequences from 82 strains of C. neoformans and C. gattii from transplant patients and 63 reference strains, covering the main molecular types described. The numbers close to the branches represent indices of support (maximum likelihood/neighbor-joining) based on 1000 bootstrap replications. The branches with bootstrap support higher than 70% are indicated in bold.
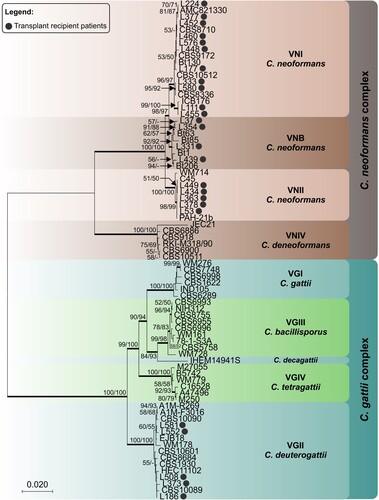
Figure 3. Median-joining haplotype network of 144 isolates of C. neoformans/C. gattii species complexes (81 isolates originated in this study in addition to 63 reference strains recovered from literature), covering all the concatenated loci CAP59, LAC1, PLB1, SOD1, URA5, TEF1 and IGS1 sequences. The isolates are coded, and their frequencies are represented by (A) fluconazole MIC ≥ than 16 mg l−1 from transplant recipients isolates or (B) 90-days mortality of transplant recipients. The size of the circumference is proportional to the haplotype frequency. The black dots (median vectors) represent unsampled or extinct haplotypes in the population. Further information about isolate source and GenBank accession number can be found in the Supplementary Table 1.
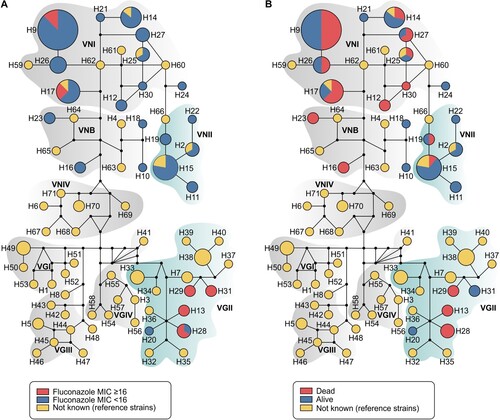
Table 2. Comparisons of infections due to C. neoformans and C. gattii complex in 60 renal transplant recipients.
Table 3. Fluconazole MIC distribution for 82 isolates of C. neoformans/C. gattii species complexes tested.
Figure 4. Kaplan–Meier analysis of 12 weeks survival of 60 renal transplant recipient infected by C. neoformans/C. gattii species complexes according to fluconazole MIC ≥ 16 mg l−1 (n = 8) or MIC < 16 mg l−1 (n = 52).
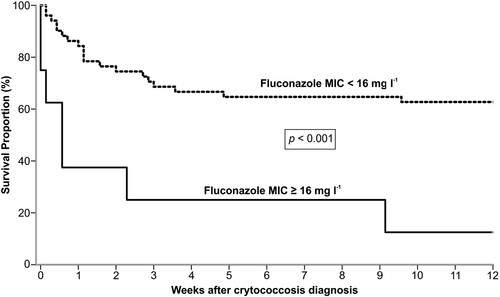
Table 4. Variables associated with 30-day mortality after cryptococcosis in 60 renal transplant recipients.
Data availability
The sequences of all newly identified allele types have been submitted to the MLST database (http://mlst.mycologylab.org) and GenBank (https://www.ncbi.nlm.nih.gov/genbank/).
