Figures & data
Figure 1. Phylogenetic and structural analysis of PRVs. (A) Phylogenetic relationship of genome sequences between PRVs isolated from human and animals. We retrieved 35 publicly available whole sequences from NCBI and partial genomes for the PRV isolated from a patient [Citation12]. Sequences were aligned by MAFFT. After getting conserved blocks from sequence alignment by Gblocks v0.91b, RAxML v8.2.4 was used to reconstruct the phylogenetic tree under the GTR+G model, with bootstrapping of 1000 replicates. iTOL v4 was used to visualized the phylogeny. The size of the circle is proportional to the bootstrap value, and only bootstrap values ≥70 was visualized. Accession numbers of PRV sequences used in this study were as follows: BK001744.1, KU360259.1, KT983811.1, KT983810.1, MG551317.1, MG551316.1, MG589642.1, NC_006151.1, LC342744.1, KX423960.1, KU900059.1, KP722022.1, KM189913.1, KM189914.3, KT824771.1, KT809429.1, KP257591.1, KP098534.1, KM061380.1, KJ789182.1, KJ717942.1, LT934125.1, KU056477.1, KU552118.1, JQ809330.1, JQ809329.1, KU315430.1, KU057086.1, KU198433.1, KM189912.1, KC981239.1, JF797219.1, JF797218.1, JF797217.1, and JQ809328.1. (B) Structure of PRV gD binding to the nectin-1 receptor. An alignment of amino acid residues in nectin-1 of various animal species are shown, and the residues important for binding (providing 5 or more Van der Waals contacts) are denoted with arrows.
![Figure 1. Phylogenetic and structural analysis of PRVs. (A) Phylogenetic relationship of genome sequences between PRVs isolated from human and animals. We retrieved 35 publicly available whole sequences from NCBI and partial genomes for the PRV isolated from a patient [Citation12]. Sequences were aligned by MAFFT. After getting conserved blocks from sequence alignment by Gblocks v0.91b, RAxML v8.2.4 was used to reconstruct the phylogenetic tree under the GTR+G model, with bootstrapping of 1000 replicates. iTOL v4 was used to visualized the phylogeny. The size of the circle is proportional to the bootstrap value, and only bootstrap values ≥70 was visualized. Accession numbers of PRV sequences used in this study were as follows: BK001744.1, KU360259.1, KT983811.1, KT983810.1, MG551317.1, MG551316.1, MG589642.1, NC_006151.1, LC342744.1, KX423960.1, KU900059.1, KP722022.1, KM189913.1, KM189914.3, KT824771.1, KT809429.1, KP257591.1, KP098534.1, KM061380.1, KJ789182.1, KJ717942.1, LT934125.1, KU056477.1, KU552118.1, JQ809330.1, JQ809329.1, KU315430.1, KU057086.1, KU198433.1, KM189912.1, KC981239.1, JF797219.1, JF797218.1, JF797217.1, and JQ809328.1. (B) Structure of PRV gD binding to the nectin-1 receptor. An alignment of amino acid residues in nectin-1 of various animal species are shown, and the residues important for binding (providing 5 or more Van der Waals contacts) are denoted with arrows.](/cms/asset/9a77b0fc-1615-4a63-83ba-c7e9bcd2059f/temi_a_1563459_f0001_oc.jpg)
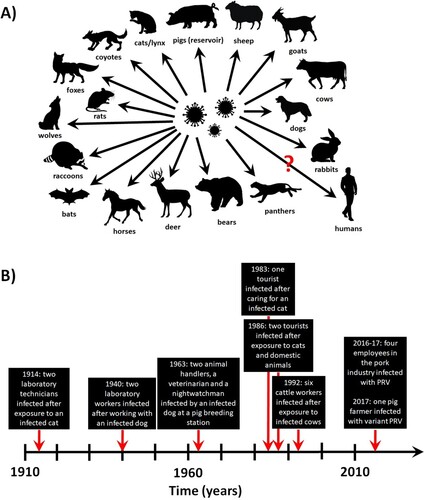
Figure 2. PRV infection in animals and humans. (A) Confirmed and potential (marked by a question mark) species susceptible to natural PRV infections. (B) Timeline of the reported PRV infections in humans.