Figures & data
Table 1. Inhibition of pathogenic fungi by cisplatin using microdilution assay.
Figure 1. Efficacy of cisplatin in a mouse model. (A) Fungal loads of Cryptococus neoformans cells in colony forming unit (CFU) were enumerated in the lungs of mice after treatment with the vehicle control (saline) and cisplatin for 4 days. Six mice per group were used. Data are expressed as mean+SD; the experiments were performed three times. (*) p<.05, (**) p<.01, and (***) p<0.001. (B) Light micrographs of Haemotoxylin and Eosin staining sections of the lungs after treatment with cisplatin. The cryptococci are indicated by arrows and are absent in the 8 mg/kg cisplatin treatment.
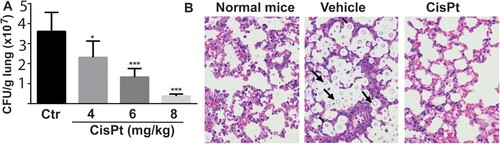
Figure 2. Cisplatin is a potent inhibitor of the Prp8 intein in vitro. (A) The Prp8 intein splicing assay based on split luciferase. RLuc (2 nM) or RLuc-Prp8 (2 nM) was used with DMF or cisplatin (40 µM) with/without 100 µM TCEP. Reactions were incubated at room temperature for 18 h, followed by luminescence detection. N=8. ***, p<.001. (B) Dose-response fitting of inhibition of splicing of the RLuc-Prp8 by cisplatin. RLuc-Prp8 (2 nM) was used. Cisplatin was in twofold serial dilutions with concentrations ranging from 100 µM (30 µg/ml) to 0.78 µM (0.23 µg/ml). N=3.
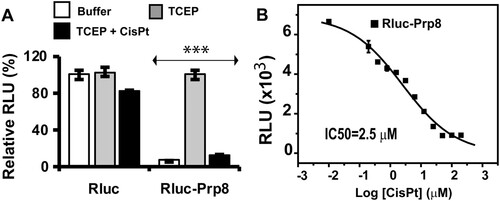
Figure 3. Cisplatin directly binds the Prp8 intein and inhibits Prp8 intein splicing. (A) MIG strategy and GFP-containing splicing products. (B) Dose-dependent inhibition of MIG-Prp8 splicing by CisPt with 100 µM TCEP. Left, SDS-PAGE analysis of each sample; right, relative percentage reduction of the MIG precursor upon treatment, compared to the starting material (T0). %P decrease is calculated as (%PT0−%PSample)/(%PT0−%PDMF)*100, where %P indicates percent of the MIG precursor at time point 0 (PT0) and/or after ∼18 h of treatment with DMF (PDMF) or cisplatin at different concentrations (PSample). N=3. **, p<.01. (C) Native-gel analysis of Prp8 (30 µM) in complex with CisPt (300 µM) in the presence or absence of 2 mM TCEP. (D) MS results of the WT Prp8 intein and its CisPt complex with/without TCEP.
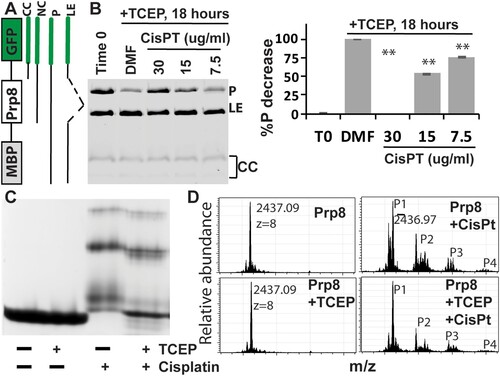
Figure 4. Crystal structure of the Cga Prp8 intein and its complex with cisplatin. (A) Overall structure of the Cga Prp8 intein. Active site residues in the conserved blocks are shown in stick-representation. (B) Superimposition of the structure of the Cga Prp8 intein with those of the Sce VMA1 and the Ssp DnaB inteins. Regions that show large structural differences between these structures are circled. N- and C-terminus are labeled. (C) Superimposition of the active site residues of the Prp8 and VMA1 inteins. (D) Structural superimposition of the native and cisplatin-bound Prp8 inteins and close-up view of interactions between the Prp8 intein and the Pt atom of cisplatin (sphere). Ligands for the platinum atom are linked with dashed lines.
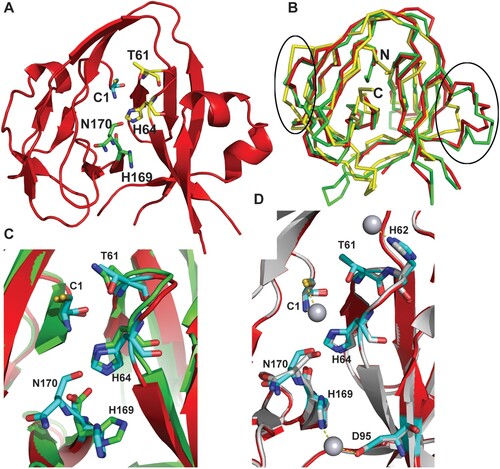
Figure 5. Alignment of sequences of intein-containing Prp8s of representative fungi known to cause human diseases with those of representative intein-free Prp8s at the intein insertion site. Conserved intein elements are in cyan. Abbreviations of organisms not defined in main text: Blastomyces dermatitidis (Bder), Botrytis cinerea (Bci), Cladophialophora bantiana (Cba), Emergomyces pasteurianus (Epas), Emmonsia parva (Epar), Exophiala oligosperma (Eol), Fonsecaea pedrosoi (Fpe), Histoplasma capsulatum (Hca), Microsporum canis (Mca), Neosartorya fischeri (Nfi), Paracoccidioides brasiliensis (Pbr), Penicillium digitatum (Pdi), Rhinocladiella mackenziei (Rma), Rhizopus microsporus (Rmi), Sporothrix brasiliensis (Sbr), Sporothrix schenckii (Ssc), Stachybotrys chartarum (Scha), Trichophyton rubrum (Tru), and chicken (Chick).

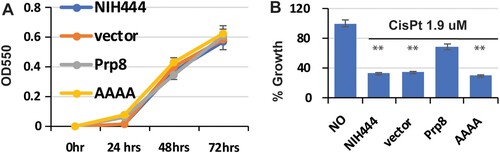
Figure 6. Overexpression of the Prp8 intein in the Cga NIH444 strain confers cisplatin resistance. (A) Transformation of NIH444 with empty vector, Prp8, and AAAA mutant does not affect the growth of NIH444. Transformed cells were cultured and monitored by OD550 at the time point indicated. N=3. (B) Transformation of the WT, but not the AAAA Prp8 mutant, into NIH444 results in resistance to cisplatin treatment. Transformed NIH444 cells were incubated with 1.9 µM cisplatin at 30°C for 48 hours. OD550 was measured. The OD550 value for untransformed NIH444 (control) in the absence of cisplatin was set as 100%. All others were expressed as a percentage of the control. N=3. **, p<.01.