Figures & data
Figure 1. Survival rates of mice after inoculation with different viruses. Female C57BL/6J mice (8–10 weeks old) were inoculated intranasally with 50 μl of PBS containing 104 TCID50 or 106 TCID50 of virus A/Yunnan-Longyang/SWL1982/2015 (YN1), A/Yunnan-Wuhua/SWL1869/2015 (YN2), A/Hunan/42443/2015 (HuN), A/Jiangsu/1/2011 (JS1), or A/Hebei-Yuhua/SWL1250/2012 (HB1). Mouse bodyweights were measured daily. Mice that lost > 30% of their original bodyweight or died naturally were recorded as “dead.”
Figure 2. Weight changes and survival rates of mice infected with HuN reassortant viruses. The pathogenicity of reassortants was evaluated by inoculating groups of mice (n = 5 per group) with 106 TCID50 (A, B) or 104 TCID50 (C, D) of virus in 50 μl of PBS or with PBS only (control). Bodyweight loss (A, C) and death (B, D) were monitored during the 14-day observation period after inoculation. rgHuN-WT, rescued wild-type HuN virus; rgHuN-PB2JS1, rescued HuN virus carrying the PB2 gene from JS1 virus; rHuN-PB1JS1, rescued HuN virus carrying the PB1 gene from JS1 virus; rgHuN-PAJS1, rescued HuN virus carrying the PA gene from JS1 virus; rgHuN-NPJS1, rescued HuN virus carrying the NP gene from JS1 virus; rgHuN-HAJS1, rescued HuN virus carrying the HA gene from JS1 virus; rgHuN-NAJS1, rescued HuN virus carrying the NA gene from JS1 virus; rgHuN-MJS1, rescued HuN virus carrying the M gene from JS1 virus; rgHuN-NSJS1, rescued HuN virus carrying the NS gene from JS1 virus.
Figure 3. Phylogenetic analysis of NP sequences of influenza A viruses and the prevalence of different amino acids at residues 305, 313, and 357. A maximum likelihood tree of NP sequences of influenza A viruses from different host species was constructed. Clusters were generally classified into Eurasian avian-like SIVs, avian influenza viruses, horse influenza viruses H3N8, human seasonal influenza viruses, 1918 H1N1 viruses, 2009 pdmH1N1 viruses, classical SIVs, and horse influenza virus H7N7 (shown from top to the bottom in ). Pie charts indicate the proportions of different amino acids at positions 305, 313, and 357 in the NP proteins in each cluster. Red triangles indicates JS1-like viruses. Red stars indicate HuN-like viruses.
Figure 4. The pathogenicity of HuN mutants in mice. Mice (n = 5 per group) were inoculated with 106 TCID50 (A, B) or 104 TCID50 (C, D) of HuN mutants. Body loss (A, C) and death (B, D) were monitored for the 14 days post-inoculation. rgHuN-WT, wild-type virus carrying NP-305K, – 313V, and – 357K; rgHuN-NPK305R, with NP-305R, – 313V, and – 357K; rgHuN-NPV313F, with NP-305K, – 313F, and – 357K; rgHuN-NPK357Q, with NP-305K, – 313V, and – 357Q; rgHuN-NPK305R+V313F, with NP-305R, – 313F, and – 357K; rgHuN-NPK305R+K357Q, with NP-305R, – 313V and – 357Q; rgHuN-NPV313F+K357Q, with NP-305K, – 313F and – 357Q; rgHuN-NPK305R+V313F+ K357Q, with NP-305R, – 313F and – 357Q.
Figure 5. Morbidity, mortality, replication, and polymerase activity assays of rgHuN-WT (357K) and rgHuN-NPK357Q (357Q) viruses in mice. Female C57BL/6J mice (8–10 weeks old) were inoculated intranasally with 50 μl of PBS containing 101 TCID50–106 TCID50 of the recombinant virus rgHuN-WT or rgHuN-NPK357Q, or with PBS (mock), to evaluate virally induced morbidity (A) and mortality (B). Morbidity was assessed by bodyweight changes over a 14-day period and was graphed as a percentage of the average weight on the day of inoculation (day 0). The average bodyweight change in each group is shown with error bars representing the standard deviations (±). Mortality associated with infection with the recombinant virus rgHuN-WT (357K) or rgHuN-NPK357Q (357Q) was also examined. (C) Viral replication efficiency in the respiratory tracts of mice was analysed by inoculating mice (n = 3 per group per time point) intranasally with 105 TCID50 of the recombinant viruses. At 1, 4, and 7 days postinoculation, three mice in each group were euthanized. Tissues, including nasal turbinate (NT), tracheal tissue (Tr), and lung (Lu), were collected for virus titration. (D) Polymerase assay was conducted in 293T cells transfected with reporter plasmids pFluc (encoding firefly luciferase flanked by the noncoding regions of the influenza NP segment to produce an artificial influenza-NP-like RNA segment), pRluc (pRL-TK, encoding Renilla luciferase, for normalization), and expression plasmids encoding PB2, PB1, PA, and NP. 357K, wild-type virus carrying NP-357K; 357Q, virus carrying NP-357Q. *p < .05; **p < .01.
Table 1 Hemagglutinin inhibition antibody test of the C57BL/6 mice inoculated with recombinant rgHuN-WT or rgHuN-NPK357Q viruses.
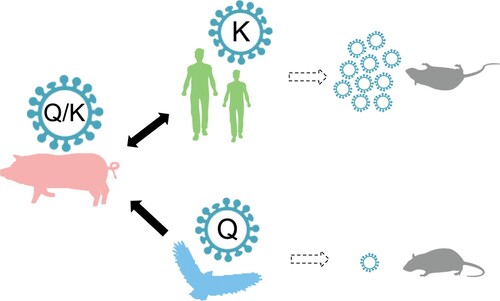
Figure 6. Conceptual model of the generalized inferred evolutionary pathways and virulence outcomes of NP-Q357K in influenza A viruses. Avian influenza viruses carrying NP-357Q were transmitted to pigs. The Q357K substitution was introduced into the NP protein during the adaptation of the avian influenza virus to swine. Mutants with NP-357K were selected, then transmitted to and circulated in humans. The Q357K substitution alters the viral phenotype in mice.