Figures & data
Table 1. Information of libraries and anogenital wart sample pools in viral metagenomic analysis.
Figure 1. Sequence read distribution of different type HPVs revealed by viral metagenomics. The counts of sequence reads aligning to different types of HPV were calculated and represented using bubbles. Each bubble represents the log 10 transformed number of HPV reads. Species and type of HPVs detected in these libraries were labelled at left side and library IDs were labelled under the corresponding column.
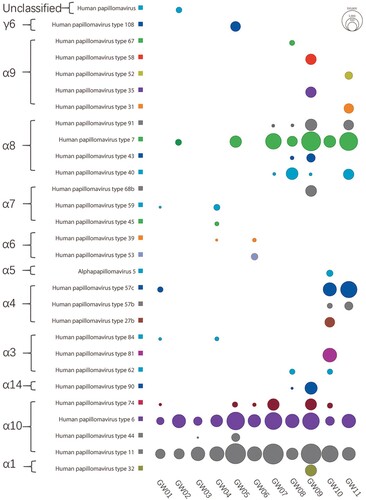
Figure 2. Phylogenetic analysis and pairwise sequence comparison based on the complete genome of HPV. The complete genomes of HPV in the phylogenetic analysis included the 24 complete genomes in this study, their closest relatives based on the BLASTn in GenBank, and other representative types of HPV in genus Alphapapillomavirus. The pairwise comparison based on complete genome among different strains within the same type of HPV was shown beside the corresponding clusters. The complete genomes acquired in this study were labelled with red dots.
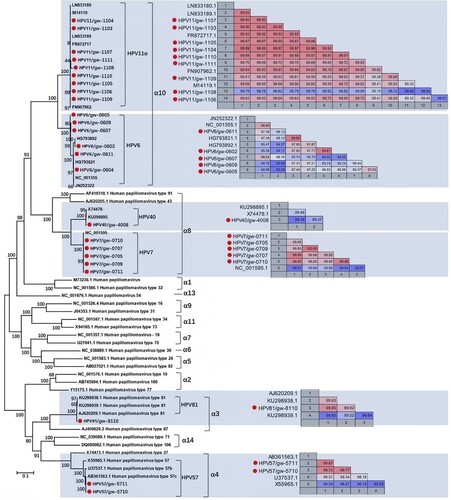
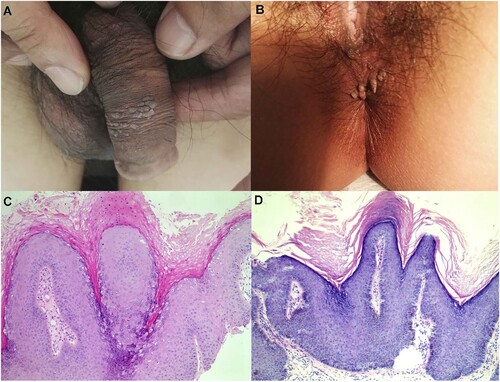
Figure 3. Representative lesions on the genital and haematoxylin and eosin staining results. (A) and (B): The two typical clinical presentation of these cases studied in this study. (C) and (D): Two representative histological findings of the two cases in (A) and (B), respectively, original magnification ×100.
Supplemental Material
Download MS Word (61 KB)Data availability
The 11 sets of raw sequence reads from the 11 viral metagenomic libraries were deposited in the Short Read Archive of GenBank database with accession no. SRX5313516- SRX5313526. The complete genome sequences human papillomavirus characterized in this study were submitted to GenBank with the accession no. MK463904-MK463926.
