Figures & data
Figure 1. Identification of host proteins associated with transcription and replication competent polymerase complex of EBOV. (A) C-terminal HA-tagged NP interacted with other viral proteins of EBOV polymerase complex. Plasmids encoding GFP-HA (negative control) or NP-HA were co-transfected separately with NP-FLAG, VP35-FLAG and VP30-FLAG into HEK293T cells. At 48 h post-transfection (p.t.), cells were collected, followed by anti-HA co-IP assay and western blotting. Representative results of 3 independent experiments are shown. (B) NP-HA rescued EBOV replication in the minigenome system. Plasmids of EBOV viral proteins (VP35, VP30, L, and empty vector, non-tagged NP or NP-HA), T7 polymerase, EBOV minigenome and pGL4 were co-transfected into HEK293T cells. At 48 h p.t., cells were collected, followed by dual-luciferase assay. The ratio of Renilla to Firefly luciferase activity of lysates from cells transfected with non-tagged NP was set to 100%. The mean and SEM from one representative experiment (n = 3) of 3 independent experiments are indicated. (C) Immunoprecipitation of host proteins associated with EBOV polymerase complex using NP-HA as bait. Plasmids expressing NP-HA, VP35, VP30, L, T7 polymerase and EBOV minigenome were co-transfected into HEK293T cells. At 48 h p.t., cells were lysed, followed by anti-HA immunoprecipitation. Eluate was separated by SDS-PAGE and visualized by silver staining. (D) Venn diagram showing the overlap of candidate host proteins interacting with NP between this study and two previous studies. Seventeen proteins that were identified both in our study and the study by Garcia-Dorival et al. are listed [Citation30].
![Figure 1. Identification of host proteins associated with transcription and replication competent polymerase complex of EBOV. (A) C-terminal HA-tagged NP interacted with other viral proteins of EBOV polymerase complex. Plasmids encoding GFP-HA (negative control) or NP-HA were co-transfected separately with NP-FLAG, VP35-FLAG and VP30-FLAG into HEK293T cells. At 48 h post-transfection (p.t.), cells were collected, followed by anti-HA co-IP assay and western blotting. Representative results of 3 independent experiments are shown. (B) NP-HA rescued EBOV replication in the minigenome system. Plasmids of EBOV viral proteins (VP35, VP30, L, and empty vector, non-tagged NP or NP-HA), T7 polymerase, EBOV minigenome and pGL4 were co-transfected into HEK293T cells. At 48 h p.t., cells were collected, followed by dual-luciferase assay. The ratio of Renilla to Firefly luciferase activity of lysates from cells transfected with non-tagged NP was set to 100%. The mean and SEM from one representative experiment (n = 3) of 3 independent experiments are indicated. (C) Immunoprecipitation of host proteins associated with EBOV polymerase complex using NP-HA as bait. Plasmids expressing NP-HA, VP35, VP30, L, T7 polymerase and EBOV minigenome were co-transfected into HEK293T cells. At 48 h p.t., cells were lysed, followed by anti-HA immunoprecipitation. Eluate was separated by SDS-PAGE and visualized by silver staining. (D) Venn diagram showing the overlap of candidate host proteins interacting with NP between this study and two previous studies. Seventeen proteins that were identified both in our study and the study by Garcia-Dorival et al. are listed [Citation30].](/cms/asset/cd044a03-5d29-4e60-95c1-a34fc44bd6d3/temi_a_1662736_f0001_oc.jpg)
Figure 2. SMYD3 was required for the replication of EBOV. (A) Depletion of SMYD3 protein expression by siRNA. HEK293T cells were transfected with negative control siRNA (siNC) or two SMYD3 specific siRNA, siSMDY3-1 and siSMYD3-2. Cells were collected 48 h p.t., followed by western blotting. Representative results of at least 3 independent experiments are shown. (B) Depletion of SMYD3 impaired the replication of EBOV in minigenome system. HEK293T cells were transfected with siNC, siSMYD3-1 or siSMYD3-2. Twelve hours later, cells were transfected plasmids for EBOV minigenome system. Cells were collected 48 h after siRNA transfection, followed by dual-luciferase assay. The ratio of Renilla to Firefly luciferase activity of lysates from cells treated with siNC was set to 100%. The mean and SEM from one representative experiment (n = 3) of 3 independent experiments are indicated. (C) Depletion of SMYD3 impaired the replication of EBOV in trVLP system. HEK293T cells were transfected with siNC, siSMYD3-1, or siSMYD3-2. Twelve hours later, cells were transfected with plasmids encoding NP, VP35, VP30, L and Tim-1. At 24 h p.t., medium was discarded and cells were infected with Ebola trVLPs. At 12 h post-infection (p.i.), supernatants were discarded and replaced with fresh medium. At 48 h p.i., cells were collected, followed by dual-luciferase assay. The ratio of Renilla to Firefly luciferase activity of lysates from cells treated with siNC was set to 100%. The mean and SEM from one representative experiment (n = 3) of 3 independent experiments are indicated. (D) Over-expression of SMYD3 promoted the replication of EBOV. HEK293T cells were transfected with empty vector or plasmid encoding FLAG-SMYD3. Twenty-four hours later, cells were transfected with plasmids for EBOV minigenome system. Cells were collected 48 h after the second transfection, followed by dual-luciferase assay and western blotting. The ratio of Renilla to Firefly luciferase activity of lysates from cells transfected with empty vector was set to 100%. The mean and SEM from one representative experiment (n = 3) of 3 independent experiments are indicated. *P < 0.05, **P < 0.01 (student’s t-test).
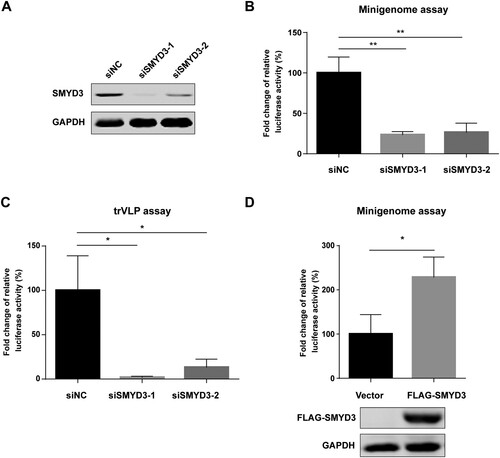
Figure 3. SMYD3 specifically interacted with EBOV NP. (A) Endogenous SMYD3 was co-purified with NP. Plasmids encoding GFP-HA, VP35-HA, VP30-HA and NP-HA were separately transfected into HEK293T cells. Cells were collected 48 h p.t., followed by anti-HA co-IP assay and western blotting. Representative results of 3 independent experiments are shown. (B) SMYD3 was recruited to EBOV inclusion bodies by NP. HEK293T cells were transfected with empty vector (panel i) or plasmids encoding NP-HA, VP35, VP30 and L (panel ii), NP-HA (panel iii), VP35-HA (panel iv) or VP30-HA (panel v). At 48 h p.t., cells were fixed with 4% poly-formaldehyde, followed by Immunofluorescence assay (IFA). Scale bar, 10 μm.
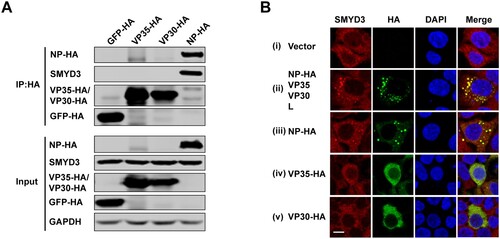
Figure 4. Mapping binding sites of NP interacting with SMYD3. (A) NP mutant with AA residues 450-600 truncated could not co-purify SMYD3. Schematic of full-length NP and 5 primary truncated mutants (top panel). Wild-type NP and mutants were separately transfected to HEK293T cells. Cells were collected 48 h p.t., followed by anti-HA co-IP assay and western blotting (bottom panel). Representative results of at least 3 independent experiments are shown. (B) Amino acid residues 556-600 of NP were essential for the interaction between NP and SMYD3. Illustration of full-length NP and deletion mutants (top panel). HEK293T cells were transfected with wild-type NP or mutants as indicated. Cells were collected 48 h p.t., followed by anti-HA co-IP assay and western blotting (bottom panel). Representative results of at least 3 independent experiments are shown. (C) Alignment of NP amino acid sequences within residues 556-600 among five EBOV species. The LxxIxE and SxPxLE motif are indicated below. (D) NP bound to SMYD3 through an SxPxLE motif. HEK293T cells were transfected with wild-type NP or single point mutants as indicated. Cells were collected 48 h p.t., followed by anti-HA co-IP assay and western blotting. Representative results of 3 independent experiments are shown. (E) The interaction between SMYD3 and NP was conserved in different EBOV species. HEK293T cells were transfected with Za-NP-WT, Za-NP-ΔSMYD3, Re-NP-WT or Re-NP-ΔSMYD3 as indicated. Cells were collected 48 h p.t., followed by anti-HA co-IP assay and western blotting. Representative results of at least 3 independent experiments are shown. (F) NP-ΔSMYD3 could not recruit SMYD3 into its inclusions. C-terminal FLAG-tagged Za-NP-WT, Za-NP-ΔSMYD3, Re-NP-WT or Re-NP-ΔSMYD3 was transfected separately with HA-SMYD3 into HEK293T cells. Cells were fixed 48 h p.t., followed by IFA. Scale bar, 10 μm.
Figure 5. Colocalization of B56α, SMYD3 and VP30 with NP-WT-, NP-ΔB56-, NP-ΔSMYD3- and NP-ΔVP30-induced inclusions. B56α-HA (A), HA-SMYD3 (B), or VP30-HA (C) were co-transfected with C-terminal FLAG-tagged NP-WT, NP-Δ56, NP-ΔSMYD3 or NP-ΔVP30 into HEK293T cells. Cells were fixed 48 h p.t., followed by IFA. Scale bar, 10 μm.
Figure 6. Mapping regions of SMYD3 binding to NP. (A) AA 2-240 of SMYD3 mediated interaction with NP. Illustration of full-length SMYD3 and truncated mutants (top panel). HA-tagged wild-type SMYD3 and truncated mutants were separately transfected with NP-FLAG into HEK293T cells. Cells were collected 48 h p.t., followed by anti-HA co-IP assay and western blotting (bottom panel). Representative results of at least 3 independent experiments are shown. (B) AA 2-240 of SMYD3 were responsible for relocation to NP-induced inclusions. N-terminal HA-tagged wild-type SMYD3 and truncated mutants were separately transfected with NP-FLAG into HEK293T cells. Cells were fixed 48 h p.t., followed by IFA. Scale bar, 10 μm.
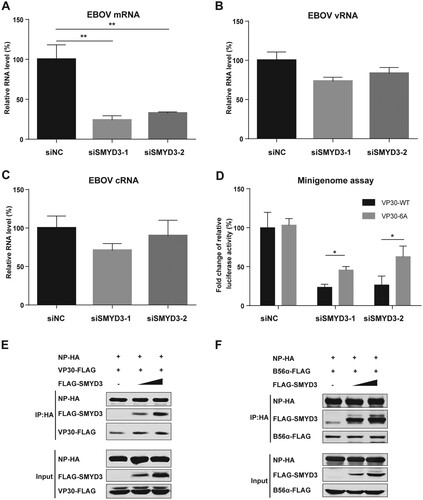
Figure 7. SMYD3 supported EBOV replication by facilitating mRNA production. (A-C) Depletion of SMYD3 down-regulated mRNA production of EBOV. HEK293T cells were transfected with siNC, siSMYD3-1 or siSMYD3-2. At 12 h p.t., plasmids for EBOV minigenome system were transfected. At 48 p.t. of siRNA, cells were collected and RNAs were extracted and analysed by qRT-PCR to measure expression levels of EBOV mRNA (A), vRNA (B), and cRNA (C). Viral mRNA, vRNA and cRNA were reverse transcribed by specific primer. qRT-PCR were performed to measure the relative RNA levels of Renilla to Firefly luciferase. The relative RNA level of cells treated with siNC were set to 100%. The mean and SD from one representative experiment (n = 3) of 3 independent experiments are indicated. (D) VP30-6A rescued replication of EBOV after depletion of SMYD3. HEK293T cells were transfected with siNC, siSMYD3-1 or siSMYD3-2. Twelve hours later, cells were transfected with plasmids of EOBV proteins (NP, VP35, L, and VP30-WT or VP30-6A), T7 polymerase and EBOV minigenome. At 48 h after transfection of siRNA, Cells were collected, followed by dual-luciferase assay. The ratio of Renilla to Firefly luciferase activity of lysates from cells transfected with siNC and VP30-WT was set to 100%. The mean and SEM from one representative experiment (n = 3) of 3 independent experiments are indicated. (E) SMYD3 promoted NP-VP30 interaction in a dose-dependent manner. HEK293T cells were transfected with NP-HA, VP30-FLAG and increasing amounts of FLAG-SMYD3. Empty vector was used to balance the total amounts of plasmids transfected. At 48 h p.t., cells were collected, followed by anti-HA co-IP assay and western blotting. Representative results of 3 independent experiments are shown. (F) SMYD3 had no effect on NP-B56 interaction. NP-HA, B56α-FLAG and increasing amounts of FLAG-SMYD3 were co-transfected into HEK293T cells. Empty vector was used to balance the total amounts of plasmids transfected. At 48 h p.t., cells were collected, followed by anti-HA co-IP assay and western blotting. Representative results of 3 independent experiments are shown. *P < 0.05, **P < 0.01 (student’s t-test).