Figures & data
Figure 1. Scheme of CRISPR-MTB. Samples were added into LY buffer with microbeads, and were vortexed/heated. Supernatants were subjected to CRISPR-MTB system. Positive fluorescent signals were captured when probes were cleaved by activated Cas12a under target sequence recognized by gRNA.
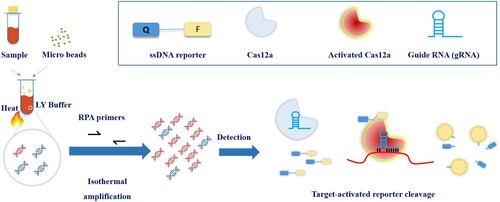
Figure 2. Performance identification of CRISPR-MTB. (a) Diameters of the beads between 0.17 and 0.11 mm are suitable for MTB broken. Each type of beads was added into lyse buffer followed same extraction and CRISPR process. No beads were served as negative control. (b) MTB DNA from optimized recipe of rapid extraction yields similar signal comparing to DNA purified by column. All three rapid extraction recipe shared same component including EDTA and Tris with indicated NaOH, CHAPS or SDS + NP40. Commercially available rapid extraction kit from Biochian was compared. DNA from column-based extraction obtained by Sangon bacteria genomic DNA purification kit was served as positive control. DNA from M. kansasii strain was rapidly extracted by SDS + NP40 and served as negative control. (c) CRISPR-MTB can detect MTB DNA close to single copy level. (d) CRISPR-MTB yields specific signal from MTB and BCG but no other DNAs. All bacteria were purified as standard manual of bacteria genomic DNA purification kit. 5 ng genomic DNA of bacteria or 50 ng hDNA was subjected to CRISPR-MTB. ** P < 0.005, **** P < 0.0001
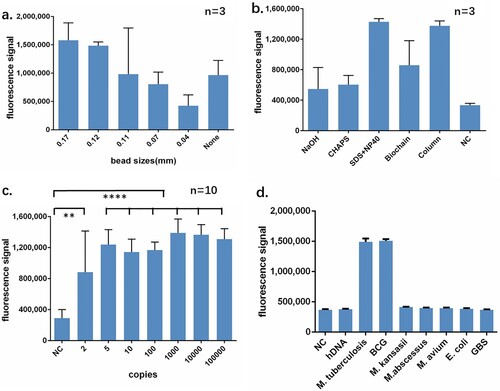
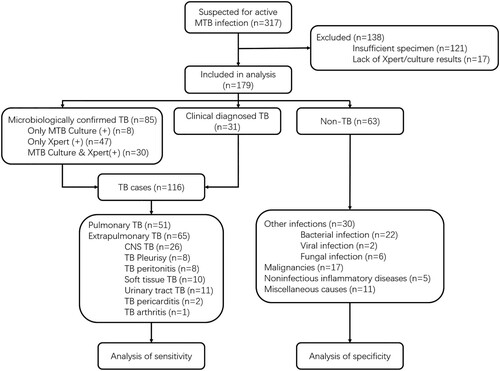
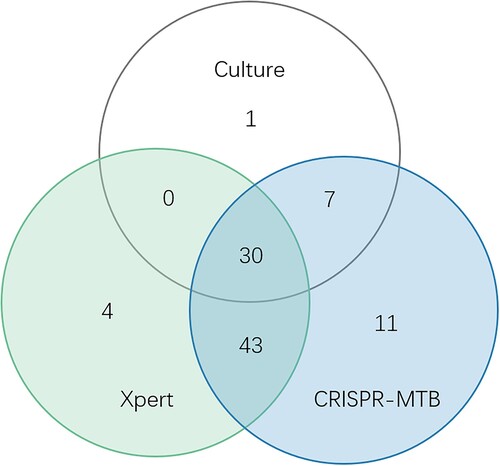
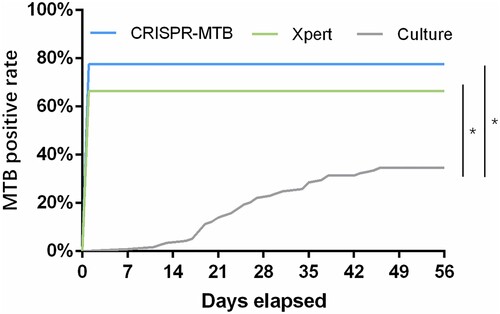
Figure 4. Cut-off value determination. (a) Typical fluorescent signal curve was presented by ABI7500 QPCR machine. (b) Fluorescent signals from each samples and positive controls were normalized by negative control of same batch. The folds changed were put together according indicated groups. “Non TB”, patients diagnosed with no active MTB infection. “Micro-confirmed TB”, patients diagnosed with active MTB infection by either culture or Xpert or both. “Clinical TB”, patients diagnosed with active MTB infection following Diagnostic criteria and principles of management of infectious MTB without evidence of either culture or GeneXpert.
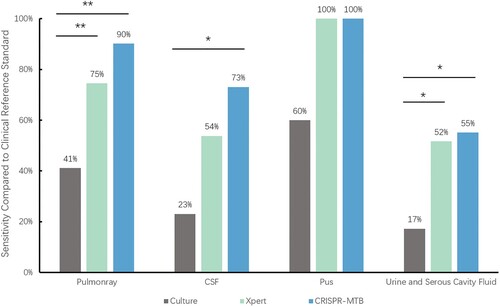
Figure 5. The sensitivity according to specimen type. * McNemar test, P < 0.01; ** McNemar test, P < 0.001