Figures & data
Figure 1. Phylogenetic reconstruction using maximum likelihood estimation of complete polyprotein of known and putative pestiviruses. Main bootstrap values are presented at nodes. Scale bar indicates number of amino acids changes per site. Taxon names are presented by the virus species and virus abbreviation. Abbreviations: CSFV, Classical swine fever virus; AydinPeV, Aydin-like pestivirus; BDV, Border disease virus; GPeV, Giraffe pestivirus; HoBiPeV, HoBi-like pestivirus; BVDV, Bovine viral diarrhea virus; PAPeV, Proghorn antelope pestivirus; PhoPeV, Phocoena pestivirus; LINDA, Lateral shaking inducing neurodegenerative agent; BuPV, Bungowannah porcine pestivirus; RPeV, Rat pestivirus; BatPeV, Bat pestivirus; APPV, Atypical porcine pestivirus
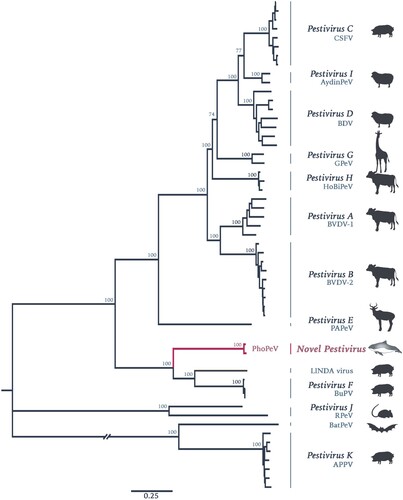
Figure 2. Genomic characterization of Phocoena pestivirus. (a) Comparison of PhoPeV to other pestivirus genome organization. PhoPeV genome arrangement is similar to other pestiviruses except for the unique absence of Npro in screened harbour porpoises (n = 7). Alignment of Npro area of PhoPeV and other pestivirus sequences is zoomed in. Absence of Npro gene coding sequences is highlighted in grey, conserved area in the capsid (from 221nt) is in bold. C/S cleavage site between Npro and Capsid is highlighted in pink and indicated with an arrowhead. Virus (Abbreviation – GenBank Accession No.): LINDA pestivirus (LINDAPeV-KY436034), Bungowannah pestivirus (BuPV-EF100713), Classical swine fever (CSFV-X87939), Aydin-like pestivirus (AydinPeV-JX428945), Border disease virus (BDV-AF037405), Giraffe pestivirus (GiPeV-AD144617), Bovine viral diarrhea virus-1 (BVDV-1-M31182), Bovine viral diarrhea virus-2 (BVDV-2-U18059), HoBi-like pestivirus (HoBiPeV-AB871953), Proghorn pestivirus (PAPeV-AY781152). (b) Comparison between PhoPeV sequences in different organs generated from two animals. The lung of animal NS170386 has an insertion (yellow) between p7 and NS2, which sequences originates from a region between NS5A and NS5B (striped yellow square). Duplicated and origin of duplicated sequence are zoomed in and are highlighted in teal colour.
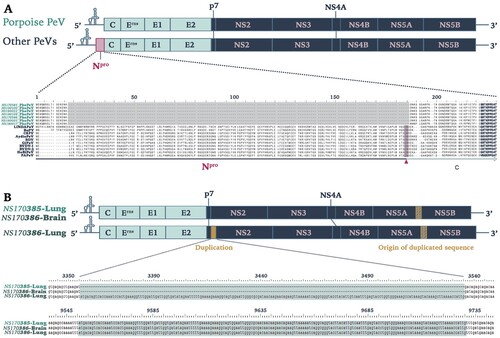
Figure 3. Phocoena pestivirus infects different cell types of harbour porpoises without histopathological changes. PhoPeV RNA expression is visible as bright red cytoplasmic staining in smooth muscle cells in the wall of an intestinal artery, epithelial cells in cortical tubules of the kidney, and neurons in the cerebrum of the brain, based on in situ hybridization (ISH) specific for Phocoena pestivirus (top row). Negative control ISH sections of these stain negative (middle row). Serial sections of these tissues, stained by hematoxylin and eosin, do not show any histopathological changes (bottow row). The narrow clefts in the neuropil of the brain are due to freeze-thaw artifact. Original objective magnifications for all panels: 40×. Artery and kidney were from porpoise NS170386, brain was from porpoise NS170385.
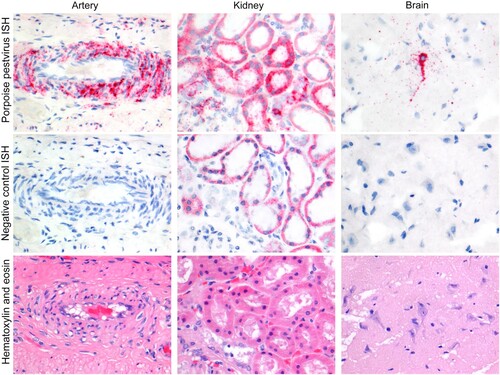
Table 1. Cell types throughout the organ systems of two harbour porpoises (NS170385-86) infected by PhoPeV. Tissue sections were stained for PhoPeV RNA by in situ hybridization.
Table 2. Lesions and levels of PhoPeV RNA (inversely correlated with Ct values) in the tissues of PhoPeV-positive harbour porpoises.
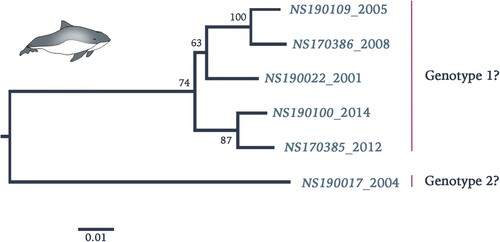
Figure 4. Phylogenetic reconstruction using maximum likelihood estimation of partial PhoPeV nucleotide genomes. Two main clades are highlighted as probable genotype 1 and 2. The PhoPeV nucleotide genomes used for analysis were 5′UTR, C, Erns, and E2 (GenBank accession nos. MK910230-37). Bootstrap values are presented at nodes. Scale bar indicates number of nucleotide changes per site LINDA virus was used as outgroup (GenBank accession number KY436034).
Supplemental Material
Download MS Word (442.4 KB)Data availability
The sequences generated in this study from two full-length PhoPeV genomes have been deposited under GenBank accession numbers (MK910227-37). Other data are available upon request.
