Figures & data
Figure 1. Basic statistics of isolates studied. (A) Distribution of isolates by year and vaccination status. A total of 154 B. pertussis isolates were available from the Reference laboratory. The graph shows their distribution by year and the proportion by the age of patients during 2008–2016. (B) The distribution of selected isolates and their ACV antigen gene profile results from different provinces. P1: Eastern-Azarbayjan, P4: Esfahan, P8: Tehran, P9: Charmahal, P27: Mazandaran. Numbers in the parentheses represent the number of selected isolates out of the total number of isolates available for each province.
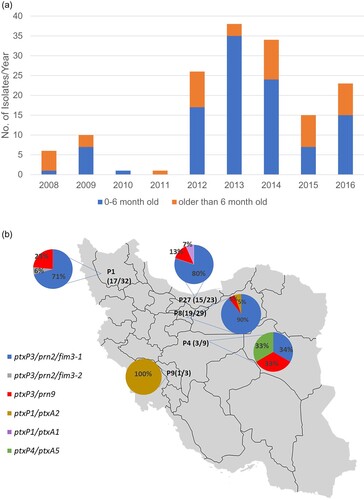
Figure 2. Maximum parsimony tree of the 54 Iranian B. pertussis isolates. The tree was constructed based on 653 SNPs. The number on the internal and terminal branches corresponds to the number of SNPs supporting each branch. Iranian ptxP3/ fim3-2 isolates were grouped into 8 clades as marked. The majority of ptxP3 isolates collected from Tehran grouped together (12 out of 18) in clade 8. New profile of ptxP3/ ptxA1/prn9 was defined as clade 5. Black circle on the tree represents the distribution of isolates collected from Tehran.
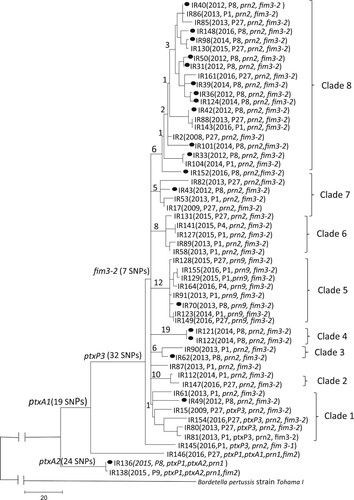
Table 1. Single nucleotide polymorphism (SNP) and short insertion and deletions (indels) supporting the nodes and clades in phylogenetic analysis.
Table 2. Unique non-synonymous SNPs in virulence associated genes or nonsense SNPs that were only found in Iranian B. pertussis isolates.
Figure 3. Phylogenetic relationships of Iranian ptxP3 B. pertussis isolates with global isolates. The tree was inferred using maximum parsimony method using B. pertussis Tohama I as an outgroup. A total of 304 ptxP3 isolates from different continents were included (Red: Iran, Blue: The USA, Green: UK, Pink: Australia) (details of the isolates are available in Supplementary File-1). Iranian isolates showed the same clustering as clade 2–8 except isolates in clade 1 that were interspersed with isolates from other countries. Clade 5 had 12 clade supporting SNPs, seven of which were shared with UK98 prn2 isolate (green circle) and two nsSNPs positioned in BP2548 and BP3694 and one mutation in the promoter of BP0359 were still unique to Iranian isolates (Node marked with a). For 6 clade 8 specific SNPs, five were shared with one Canadian isolate (B062, isolated in 2001) and one nsSNPs located in B1245 was still unique to Iranian isolates (Node marked with b). The only ptxP3/fim3-1 Iranian isolate (IR145) was grouped with fim3-1 isolates from USA (Node marked with c).
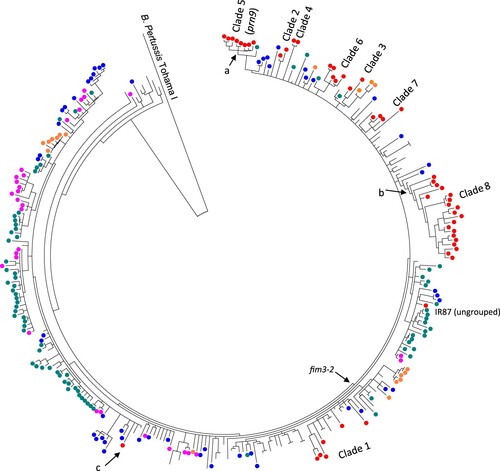
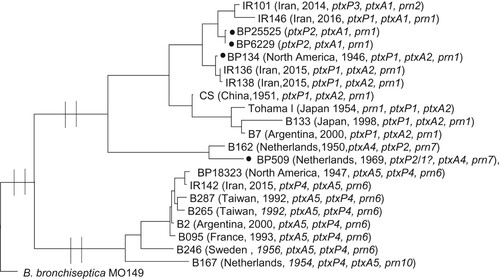
Figure 4. Maximum Parsimony Phylogeny using 2792 SNPs to analyse the relationship between old B. pertussis isolates, vaccine seed isolates and recent isolates from Iran. B. bronchiseptica MO149 used as an outgroup. Isolated marked with black circle are vaccine seeds used in WCV manufactured by Serum Institute of India that is used for immunization in Iran. BP509 and BP134 were also used in WCV in the years when Iran manufactured and used its own vaccine.