Figures & data
Figure 1. The ΔfexA mutant exhibit growth defect and severe deficiencies in acid tolerance, reactive oxygen resistance and in vivo survival. (A) Bacterial growth under aerobic, microaerobic and anaerobic conditions was determined. (B) Acid tolerance under 10 mM sodium citrate (pH 5.0) was determined as percent survival. (C) Bacterial survival in 2.5 HI broth containing 1 mM H2O2 was determined. (D) Intra-intestinal survival, growth and subsequent invasion into blood stream of V. vulnificus strains were determined. (E) Survival of mice intraperitoneally infected with the V. vulnificus strains were determined (n = 17 ∼18). The error bars represent standard errors. *P < 0.05; **P < 0.01, ***p < 0.001.
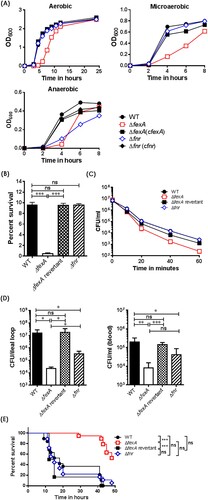
Figure 2. Differentially expressed genes of the ΔfexA strain under aerobic, anaerobic and in vivo conditions were identified by DNA microarray analysis. Venn diagram showing the extent of overlapping genes that are differentially upregulated or downregulated in the ΔfexA mutant among aerobic, anaerobic and in vivo growth conditions. Downregulated genes of the ΔfexA strain under aerobic, anaerobic and in vivo conditions.
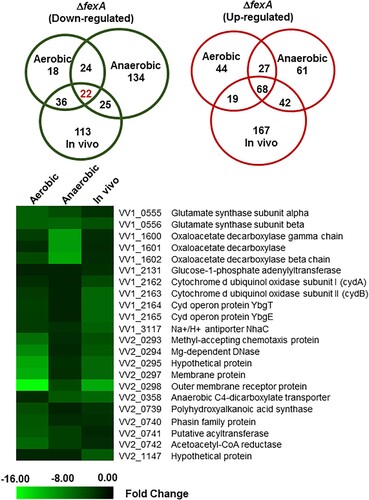
Table 1. Genes down-regulated in ΔfexA strain under aerobic, anaerobic and in vivo conditions (cutoff: fold change 2.0).
Figure 3. Compensatory mutations in the cydAB promoter region significantly enhanced cydAB promoter activity. The cydAB promoter activities were determined by measuring β-galactosidase activity of the cydAB-lacZ reporter plasmids under aerobic, microaerobic and anaerobic growth conditions. The error bars represent standard errors. ***p < 0.001.
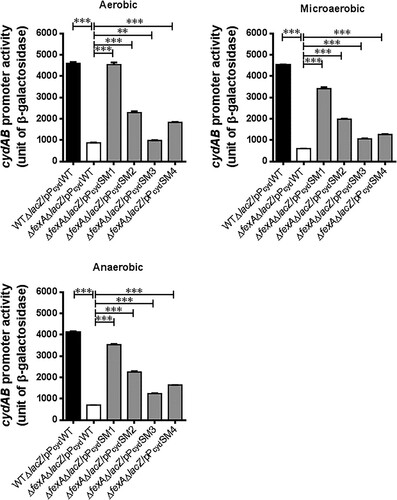
Figure 4. Compensatory mutations in the cydAB promoter region reversed phenotypic changes of the ΔfexA mutant. (A) Bacterial growth was monitored by measuring the OD600 value of cultures at different time points. Cytotoxicity (B) and motility (C) of the WT strain and respective mutants were determined. The error bars represent standard errors **p < 0.01.
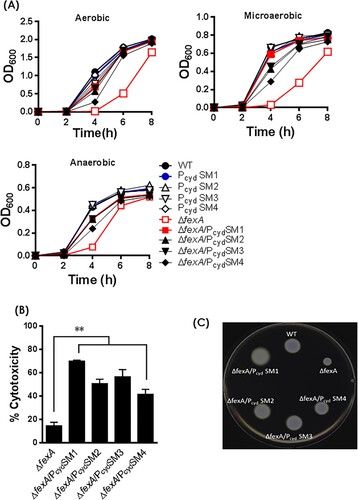
Figure 5. Compensatory mutations in the cydAB promoter region restore ATP production and the deficiencies of H2O2 resistance and in vivo survival of ΔfexA mutant. (A) Intracellular ATP production was measured under aerobic, microaerobic and anaerobic growth conditions. (B) Bacterial survival in 2.5 HI broth containing 1 mM H2O2 was determined. (C) In vivo growth of the ΔfexA mutant in the rat peritoneal cavity (n = 4) was determined by using a dialysis tube implantation model. (D) Bacterial growth in minimal essential medium (MEM) supplemented with 0.2% glucose, 0.2% glycerol or 0.2% succinate under aerobic culture condition was determined. The error bars represent standard errors. *p < 0.05; **p < 0.01; ns, not significant.
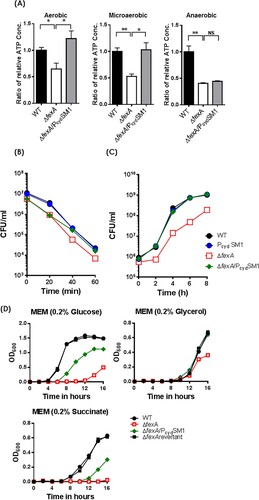
Table 2. Effect of mutations on the lethality of V. vulnificus in mice.
Figure 6. Analysis of the cydAB promoter-binding protein complex. (A) Determination of the cydAB promoter binding proteins. A biotin-labeled DNA fragment of the cydAB promoter region was affixed to streptavidin-conjugated Dynabeads, and then incubated with V. vulnificus cytoplasmic extract. Non-adhering and low-specificity DNA-binding proteins were removed by repeated washing and DNA-binding proteins were eluted. Single protein bands were cut from the SDS-PAGE gel for MALDI-TOF Mass Spectrometry assay. (B) The promoter region of the cydAB operon. Bioinformatic analysis suggests two putative FNR binding (blue colour), two putative FexA binding (red colour), two LeuO binding (highlighted), one HexR (green colour) and one SeqA (violet colour) sites.
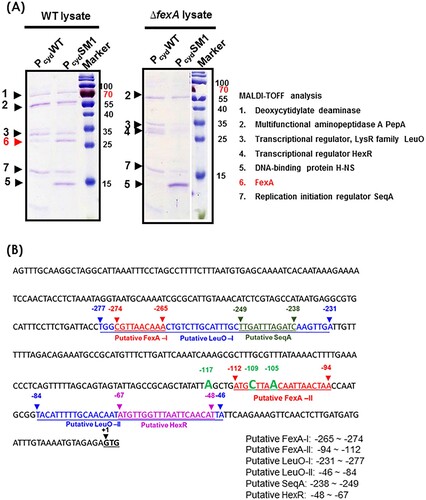
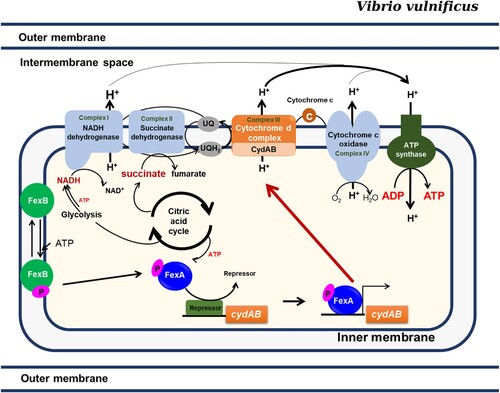
Figure 7. The FexAB two component system regulates the adaptive responses to oxygen availability. Upon stimulation, FexB undergoes autophosphorylation, and a phosphoryl group is transferred to FexA by a His-Asp-His-Asp phosphorelay, which consequently activates the expression of cydAB. CydAB encodes the terminal oxidase of the electron transport pathway, which is essential for the survival of V. vulnificus. The most important role played by FexA in V. vulnificus is the activation of cydAB expression.