Figures & data
Figure 1. GILT expression is upregulated by IFN-γ in lung cancer cells and has a lysosomal distribution. A549 and MRC-5 cells were treated with the indicated concentrations of IFN-α or IFN-γ for 12 h. (A) The levels of intracellular GILT and IFITM proteins were determined by Western blot assay. β-actin served as a loading control. (B) The localization of GILT in MRC-5 cells treated with IFN-γ was detected by immunofluorescent staining with anti-GILT polyclonal antibody (green). EEA1, Rab9 or LAMP1 were visualized by immunofluorescent staining with respective antibodies (red). Cell nuclei were stained blue with DAPI.
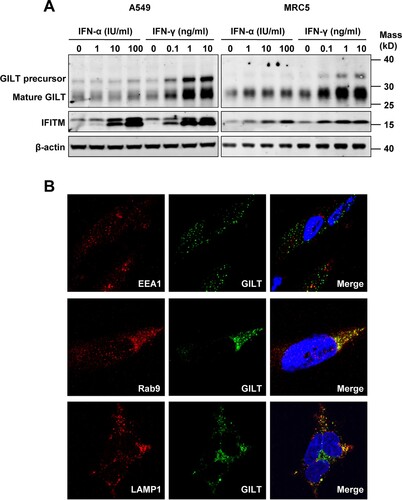
Figure 2. SARS-CoV, EBOV and LASV envelope glycoproteins-mediated infection is restricted by GILT. (A to C) FLIP-IN T Rex 293 cells that tetracycline (tet) inducibly express chloramphenicol acetyltransferase (CAT), GILT or FLAG-tagged IFITM3 proteins were cultured in the presence or absence of 1 μg/ml of tet for 24 h. The induced expression of GILT was detected by Western bot assay (A) and visualized by immunofluorescent staining (B). (C) The above-mentioned cells were then infected with indicated pseudoviruses. Pseudovirus infection was measured by luciferase assay at 48 h post infection (hpi). Relative infection represents the ratio of luciferase activity in cells treated with tetracycline over that in cells cultured without tetracycline. GILT significantly (**p < 0.001) inhibits the infection of SARSpp, but do not affect the infection of all other tested pseudoviruses. (D) The intracellular expression of GILT in A549 cell lines stably transduced with GILT or empty vector (pQCXIP) treated with IFN-γ at indicated concentration for 12 h was determined by a Western blot assay. (E) The entry mediated by GP proteins from SARS-CoV, EBOV, LASV and H1N1 in IFN-γ-treated A549 cells was determined by using the BlaM-Vpr assay. Cells were analysed by flow cytometry 24 h after infection and infectivity was determined by measuring cleaved CCF2 (Blue) from uncleaved CCF2 (Green). Relative infection efficiency was calculated by setting the infection of empty vector (pQCXIP) to 1.0. GILT significantly (**p < 0.001) inhibits the infection of SARSpp, EBOVpp and LASVpp, but do not affect the infection of IAVpp. Data are shown as the mean ± SD (standard deviation) of three independent experiments. *p < 0.05; **p < 0.001.
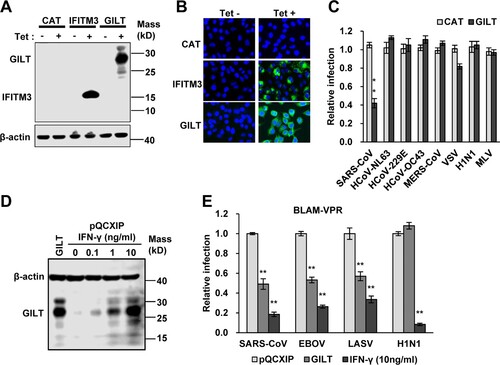
Figure 3. Knockout of GILT in THP-1 cells enhances viral entry of EBOV and LASV. (A) sgRNA sequences used in this study. (B) Schematic illustration of seven exons of GILT gene and guide RNA target regions. (C) Genomic DNAs from wild-type parental THP-1 (GILT WT) cells, GILT/KO THP-1 (GILT/KO1-1, 1–2 and GILT/KO2-1, 2–2) cells and empty vector control (CTRL) were isolated. gRNA-targeted regions in THP-1 chromosome were sequenced and aligned with GILT RefSeq. (NM_006332). (D) Cell lysates from indicated PMA-stimulated THP-1 clones were probed with a monoclonal anti-GILT antibody. β-actin severed as a loading control. (E) The above-mentioned THP-1 cells were infected with EBOVpp, LASVpp, or IAVpp, respectively. Pseudovirus infection was measured by luciferase assay at 48 hpi. Relative infection represents the luciferase activity normalized to that of THP-1 CTRL cells. Differences in relative pseudovirus infection between GILT/KO cells and THP-1 CTRL cells were statistically analysed (**p < 0.001). Data are shown as the mean ± SD of three independent experiments. *p < 0.05; **p < 0.001.
Figure 4. Mutations of N-linked glycosylation sites compromise GILT activity of inhibiting viral entry mediated by entry proteins from EBOV and LASV. (A) Illustration of GILT with three N-linked glycosylation sites and its mutants. (B) FLIP-IN T Rex 293 cells expressing GILT or indicated mutant GILT proteins were cultured in the presence of 1 μg/ml of tet for 24 h. The induced expression of indicated GILT mutants was detected by Western bot assay using anti-GILT polyclonal antibody, which recognizes both precursor and mature form. β-actin served as a loading control. (C) The above-described cell lines were infected with SARSpp, EBOVpp, LASVpp or IAVpp. Luciferase activities were determined at 48 hpi. Relative infection efficiency represents the luciferase activity in cells cultured with tetracycline normalized to that in cells cultured without tetracycline. Data are shown as the mean ± SD of three independent experiments. *p < 0.05, **p < 0.001 compared to wild-type GILT.
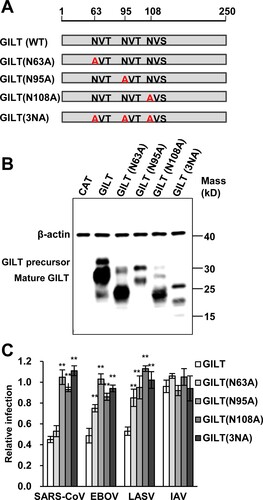
Figure 5. Mutations of N-linked glycosylation reduce GILT lysosomal localization. FLP-IN T Rex cells expressing the indicated wild-type and N-linked glycosylation mutant GILT proteins were treated with Tet for 24 h to induce GILT expression. The localization of wild-type GILT and its N-linked glycosylation mutants was detected by immunofluorescent staining with anti-GILT polyclonal antibody (green). LAMP1 was visualized by immunofluorescent staining with anti-LAMP1 monoclonal antibody (red). Cell nuclei were stained blue with DAPI (blue). More than 10 fields were examined, and the representative images of one or a few cells are presented.
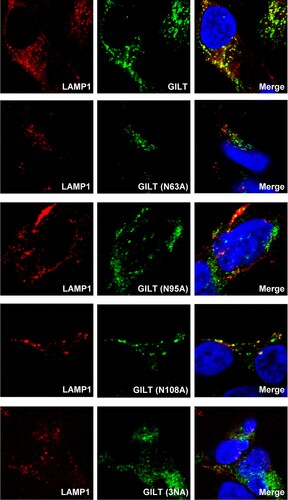
Figure 6. Cysteine mutations in thiol reductase active site compromise GILT activity to inhibit viral entry of EBOV and LASV. (A) Schematic representation of the GILT protein structural domains, motif of enzyme active centre, and N-linked glycosylation sites. (B) FLIP-IN T Rex 293 cells expressing GILT or indicated mutant GILT proteins were cultured in the presence or absence of 1 μg/ml of tet for 24 h. The expression of indicated GILT mutants was detected by Western bot assay using anti-GILT polyclonal antibody. β-actin served as a loading control. (C) The above-mentioned FLIP-IN T Rex 293 cells were infected with SARSpp, LASVpp, EBOVpp or IAVpp. Luciferase activities were examined at 48 hpi. Relative infection efficiency is the ratio of luciferase activity in cells cultured with tetracycline over that in cells cultured without tetracycline. Results are the means ± SD of three independent experiments. **p < 0.001 compared to wild-type GILT. (D) FLIP-IN T Rex 293 cells expressing wild type or mutant GILT proteins were left untreated or treated with tet for 24 h to induce the GILT expression. The expression of cathepsins L, B and S in FLIP-IN T Rex 293 cells were determined by western blot assay. (E) The cathepsin proteolytic activity in cells used in (D) were measured and presented as relative fluorescence unit. Results are the means ± SD of three independent experiments. **p < 0.001.
Figure 7. Trypsin treatment bypasses GILT restriction of entry mediated by SARS-CoV S protein. FLIP-IN T Rex 293 cells inducibly expressing IFITM3 (A) or GILT (B) proteins were left untreated or treated with tet for 24 h to induce the IFITM3 or GILT expression. Cells were then incubated with SARSpp at 4°C for 30 min and subsequently treated with TPCK-treated trypsin (5 mg/ml) or DMEM at 37°C for 15 min. The infected cells were maintained in complete DMEM and pseudovirus infection was determined by luciferase assay at 48 hpi. Relative infection represents the ratio of luciferase activity in cells treated with tetracycline over that in cells untreated with tetracycline. Results are the means ± SD of three independent experiments. Both IFITM and both GILT proteins significantly (**p < 0.001) inhibited infection by SARSpp.
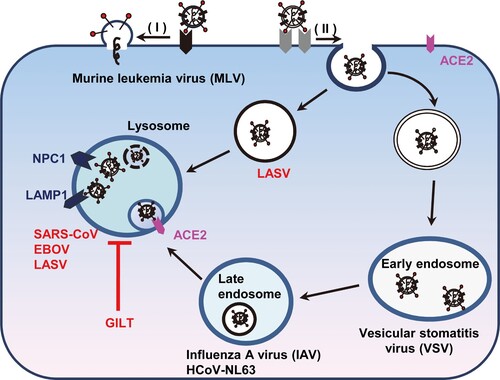
Figure 8. Schematic model for GILT-mediated restriction of viral entry in lysosome. Enveloped viruses enter cell at plasma membrane or escape from endocytic compartments via virus-host cell membrane fusion. While MLV that fuses at plasma membrane and vesicular stomatitis virus (VSV) and influenza A virus (IAV) that fuse at early endosome or late endosome are resistant to GILT-mediated inhibition, viruses including Ebola virus, Lassa virus and SARS-CoV, which fuse at lysosome, are restricted by GILT. NPC1: Niemann-Pick C1 protein (lysosomal receptor of Ebola virus); LAMP1: Lysosome-associated membrane glycoprotein 1 (lysosomal receptor of Lassa virus).