Figures & data
Figure 1. PrrH suppresses pyocyanin synthesis of P. aeruginosa. (A) PrrH overexpression inhibited pyocyanin production. PAO1 carrying the pROp200 vector (Vector) or pROp200-prrH (PrrH) was cultured in LB for 24 h, and pyocyanin in the supernatant was measured. (B) Deficiency in prrH resulted in an increase in pyocyanin, and complementation with prrH abolished pyocyanin production. PAO1 and PAO1ΔprrH carrying pROp200 (WT/Vector, ΔprrH/Vector) or pROp200-prrH (ΔprrH/PrrH) were cultured in LB for 20 h, and pyocyanin in the supernatant was measured. (C, D) Pyocyanin production was negatively correlated with PrrH expression under the treatment of AZM. PAO1 was treated with 2, 8 or 32 μg/mL of AZM for 24 h, followed by pyocyanin measurement (C) or real-time PCR analysis (D). The rpoD gene was used as an internal control. Values are the mean ± SD of at least three independent experiments. ns, not significant; **, P < 0.01; ***, P < 0.001.
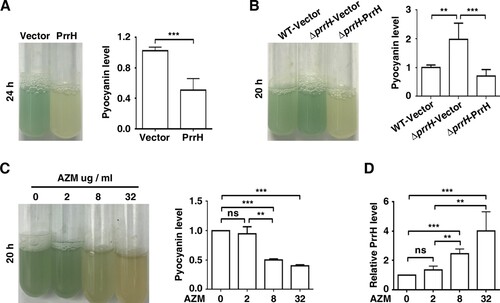
Figure 2. PrrH influences elastase and rhamnolipid production, biofilm formation, swimming and swarming motility of P. aeruginosa. (A) PrrH overexpression inhibited elastase production. PAO1 carrying pROp200 (WT/Vector), PAO1ΔprrH carrying pROp200 or pROp200-prrH (ΔprrH/Vector, ΔprrH/PrrH) were cultured in LB for 8 h, and the activity of elastase in supernatant was determined. (B) PrrH overexpression inhibited rhamnolipid synthesis. The indicated PAO1 strains were cultured in M9 minimal salts medium for 8 h at 37 °C, and rhamnolipid in the supernatant was measured. (C) PrrH inhibited biofilm formation. The indicated PAO1 strains were cultured in LB in the 12-well plates for 24 h at 37°C. The biofilm was quantified by measuring solubilized crystal violet staining biofilm cells at OD600. (D) PrrH positively regulated swimming and swarming motility. 5μl cultures of the indicated strains were spotted onto the swarming or swimming medium and incubated at 37°C for 16 h. Values are the mean ± SD of at least three independent experiments. ns, not significant; **, P < 0.01; ***, P < 0.001.
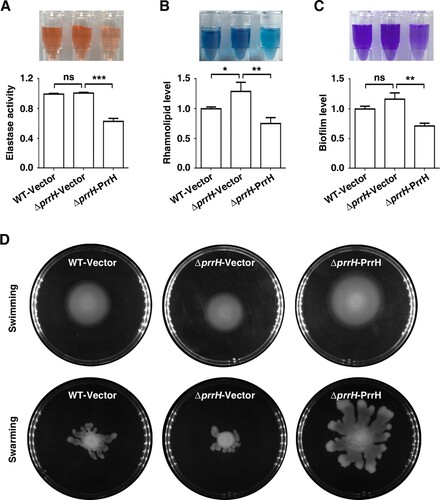
Figure 3. PrrH inhibits P. aeruginosa bacteraemia in vitro. (A) PrrH repressed P. aeruginosa survival in whole blood. Here, 1.0 × 106 CFU of wild-type PAO1 (WT/Vector) and prrH-overexpression (WT/PrrH) or prrH-deficient (ΔprrH/Vector) strains were cultured in whole blood for 30, 60, 90 and 120 min. ***P <0.001, compared with the wild-type strain. (B) The expression of PrrH was enhanced by whole blood. PAO1 was cultured in LB with or without 10% whole blood (indicated as ‘‘+’’ and ‘‘-’’, respectively) for 4 or 8 h, followed by real-time PCR analysis. The rpoD gene was used as an internal control. Values are the mean ± SD of at least three independent experiments; **, P < 0.01; ***, P < 0.001.
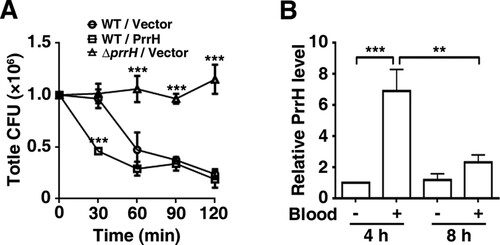
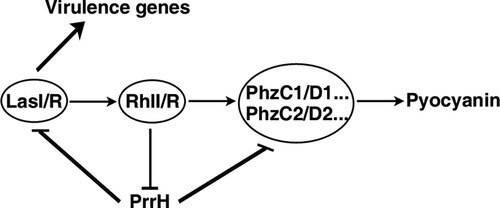
Figure 4. LasI, PhzC and PhzD are direct targets of PrrH. (A) PrrH and its putative binding sequence in the CDS of LasI, PhzC and PhzD. Asterisks denote the mutant PrrH-binding site that was generated as the complementary sequence. (B) Principles of the in vivo investigation of direct interactions between sRNAs and their targets in the E. coli DH5α strain. (C, D) PrrH suppressed the intensity of GFP through its binding sequences at the CDS of target genes. DH5α strains were co-transformed with pROp200 (Vector) or pROp200-prrH (PrrH) and a GFP reporter containing a wild-type or mutant CDS of target genes (indicated as WT or Mut); GFP was observed by fluorescence microscopy (C), and the intensity was measured by a Tecan Infinity M1000Pro Reader and expressed in AU as F485/535/Abs595 (D). ***P < 0.001, compared with pROp200-transformants. (E) Effect of prrH overexpression or deficiency on the endogenous level of LasI, LasA, PhzC and PhzD. PAO1 and PAO1ΔprrH carrying pROp200 (WT/Vector, ΔprrH/Vector) or pROp200-prrH (WT/PrrH, ΔprrH/PrrH) were cultured in LB for 6 h before qPCR analysis, and the rpoD gene was used as an internal control. Values are the mean ± SD of at least three independent experiments; **, P < 0.01; ***, P < 0.001.
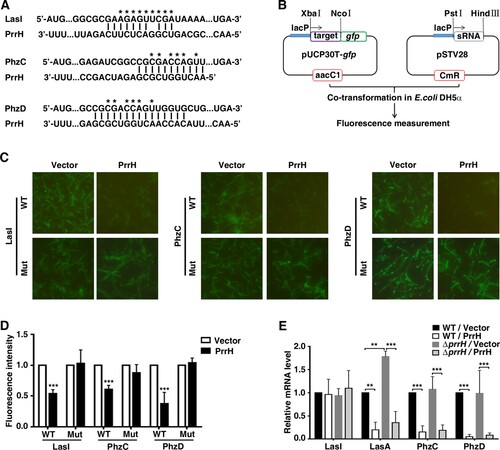
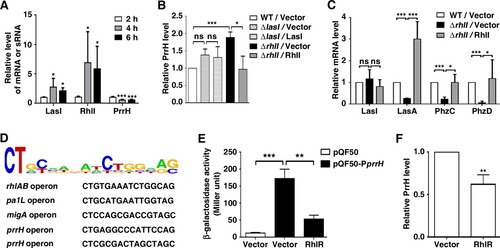
Figure 5. PrrH is negatively regulated by RhlI/R. (A) The expression of PrrH was negatively related to LasI and RhlI. PAO1 in the exponential phase was collected and re-cultured in LB for 2, 4 or 6 h, followed by qPCR analysis. (B) RhlI repressed the expression of PrrH. (C) The target genes of PrrH were regulated by RhlI. (B, C) PAO1 (WT) and the lasI- or rhlI-deficient strains (ΔlasI, ΔrhlI) carrying pROp200 (Vector), pROp200-lasI (LasI) or pROp200-rhlI (RhlI) were cultured in LB for 6 h before qPCR analysis, and the rpoD gene was used as an internal control. (D) Diagram of RhlR-binding sites in the promoter of its putative target genes. (E) Overexpression of RhlR decreased prrH promoter activity. The BW25113 strains carrying the reporter pQF50 or pQF50-PprrH combined with pROp200 or pROp200-rhlR were grown to mid-log phase and subjected to a β-galactosidase activity assay. (F) RhlR repressed the expression of PrrH. The indicated PAO1 strains were cultured for 6 h, followed by real-time PCR analysis. Values are the mean ± SD of at least three independent experiments. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.