Figures & data
Figure 1. Scheme of the reactions catalyzed by two GAR transformylases, PurT and PurN. In the third step of the de novo purine nucleotide biosynthesis pathway, PurT uses formate and ATP to catalyze GAR to N-formyl-GAR (FGAR), while PurN uses 10-formyltetrahydrofolate in this reaction.
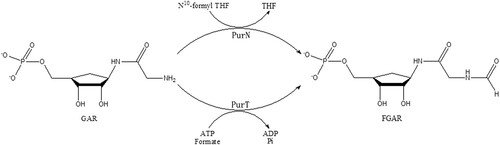
Table 1. Data collection and refinement statistics for MtbPurT.
Table 2. The primers used for amplification of MtbPurT mutants.
Figure 2. Overall structure of MtbPurT. (A) Analytical gel-filtration chromatography of final purified MtbPurT. A single peak was observed with molecular weight corresponding to the size of a dimer. The SDS-PAGE analysis of the final purified MtbPurT was shown as upper insert. (B) Ribbon representation of the MtbPurT dimer. The monomers are coloured yellow and cyan. (C) Subdomain structure of MtbPurT. MtbPurT structure comprises three subdomains, an A-domain at the N-terminus (coloured blue and labeled N), a B-domain in the middle (coloured red), and a C-domain at the C-terminus (coloured green and labeled C). The secondary structure elements are also labeled. (D) Surface representation of MtbPurT. The possible ATP and substrate binding sites are marked.
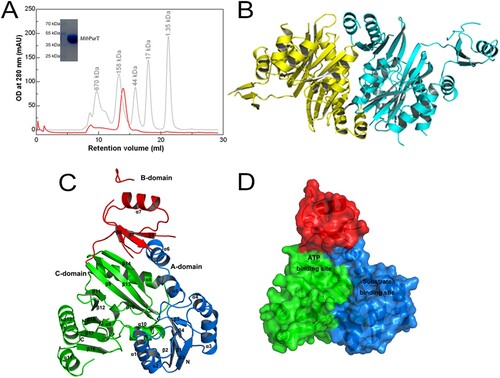
Figure 3. Structural comparison of MtbPurT with apo-phBCCPPurT. (A) Superposition of MtbPurT with apo-phBCCPPurT (PDB code: 2CZG, r.m.s.d: 1.30 Å). The shift of the loop in MtbPurT from the positions in phBCCPPurT is marked as a dashed arrow in red. (B) Surface representation of the molecular surfaces in MtbPurT and phBCCPPurT. (C) Isothermal titration calorimetry assay (ITC) demonstrates the ATP-binding affinity for MtbPurT and phBCCPPurT. The calculated Kd values are shown. (D) No binding affinity with CTP for MtbPurT was detected by ITC.
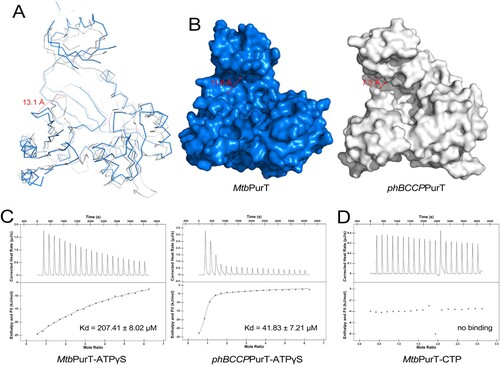
Figure 4. Structural comparison of MtbPurT with the ATP-grasp fold proteins. Structure of MtbPurT (coloured blue) was superposed with (A) phBCCPPurT-ADP (coloured yellow, PDB code: 2DWC, r.m.s.d: 1.31 Å), (B) FtPurK-AMPPNP (coloured green, PDB code: 4MA5, r.m.s.d: 1.80 Å) and (C) LlPC-ADP (coloured light purple, PDB code: 5VYZ, r.m.s.d: 2.44 Å). In each ATP-grasp fold protein, residues involved in cofactor binding are shown in sticks and coloured yellow, green and light purple, respectively. The corresponding residues in MtbPurT are presented in sticks and coloured in magenta. (D) Multiple sequence alignment of MtbPurT, phBCCPPurT, FtPurK and LlPC. Residues involved in binding with ATP-analogs are marked as triangles.
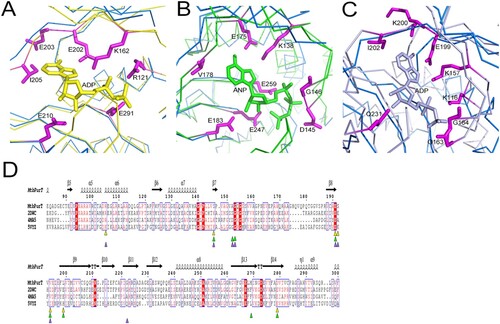
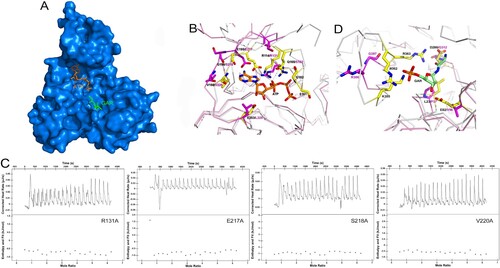
Figure 5. Model of ATP and GAR binding to MtbPurT. (A) Surface representation of MtbPurT with its proposed ATP and GAR. The ATP and GAR molecules are shown in stick representation. (B) The proposed models with ATP are displayed. Residues involved in ATP binding in EcPurT are coloured yellow, and their corresponding residues in MtbPurT are coloured magenta. (C) The ATP-binding affinity for MtbPurT mutants was measured by ITC. None of the mutants bound ATPγS. (D) The proposed models with GAR are displayed. Residues involved in GAR binding in EcPurT are coloured yellow, and their corresponding residues in MtbPurT are coloured magenta. All residues are shown in stick representations.