Figures & data
Table 1. Characteristics of mechanisms of resistance and modifications associated with polymyxin resistance.
Table 2. Main characteristics of mcr genes related to polymyxin resistance.
![Figure 1. (a) Structures of colistin A and B; (b) structures of sodium colistin A and B methanesulphonate. Fatty acid: 6-methyl-octanoic acid for colistin A and 6-methyl-heptanoic acid for colistin B; Thr: threonine; Leu: leucine; Dab: α, γ-diaminobutyric acid. α and γ indicate the respective amino groups involved in the peptide linkage. Adapted from Li et al. [Citation16].](/cms/asset/a8caac10-f439-401c-98ed-20bd0057ced8/temi_a_1754133_f0001_ob.jpg)
![Figure 2. PRISMA-modified flow diagram of included and excluded studies. Adapted from the PRISMA website (http://www.prisma-statement.org/PRISMAStatement/FlowDiagram) and Liberati et al. [Citation11].](/cms/asset/9314c283-eac9-426a-9ede-30d2322fa3ac/temi_a_1754133_f0002_ob.jpg)
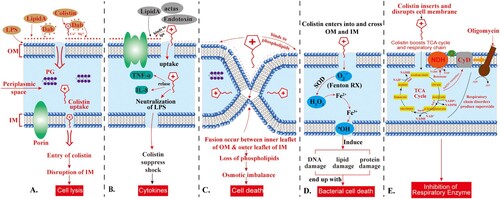
![Figure 4. Scheme of colistin binding to lipid A. (A) a Schematic of the transfer of phosphoethanolamine to the 1-PO4 group of Hexa-acylated lipid A in the presence of MCR-1. (B) Models of colistin (blue sticks) binding to lipid A (left) or phosphoethanolamine-1΄-lipid A (right) (spheres coloured green, red, blue, and orange for C, O, N, and P atoms, respectively). a (left), The positively charged Dab colistin residues interact with the negatively-charged 1′ and 4′ phosphate groups of lipid A, reducing the net-negative charge of lipid A. The hydrophobic leucine residues and tail of colistin A bind with the fatty acid tails of lipid A, allowing the uptake of colistin A, and disrupt, the bacterial OM. b (right), a model of colistin binding to phosphoethanolamine-1΄-lipid A indicates the addition of positively charged phosphoethanolamine onto the 1′-PO4 of lipid A likely interferes with the interaction of positively charged Dab8 and Dab9 side chains with the phosphate group, preventing colistin binding to the outer membrane of GNB. The model B is adapted from Yang et al. [Citation74].](/cms/asset/cad8097c-8280-4964-87f9-cf2bae7fa4a5/temi_a_1754133_f0004_oc.jpg)
