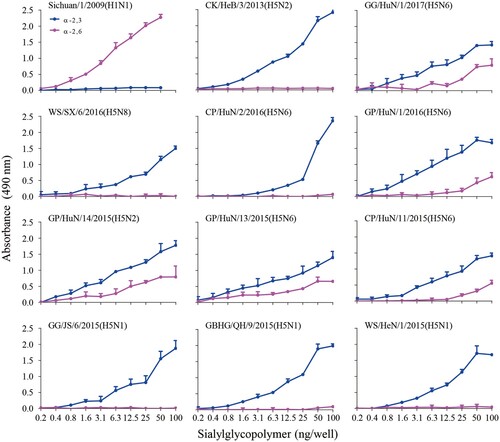
Figures & data
Figure 1. Geographical locations and bird species involved in the wild bird outbreaks caused by different genotypes of H5 viruses.
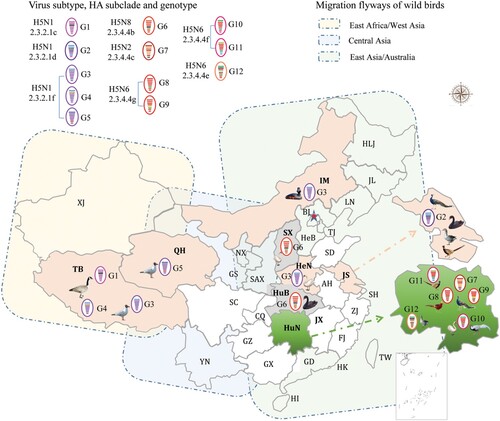
Table 1. H5 avian influenza outbreaks in wild birds in China.
Figure 2. Phylogenetic analysis of H5 wild bird viruses and genotypes. The Maximum Clade Credibility (MCC) tree of the HA gene was constructed by using the package BEAST (v1.8.4). The clades shown in different colours in the tree were annotated by using FigTree (v1.4.4) (a), and the origins of each gene segment are indicated by different coloured bars (b). Phylogenetic trees with complete information are provided as Supporting Figures S1 and S2.
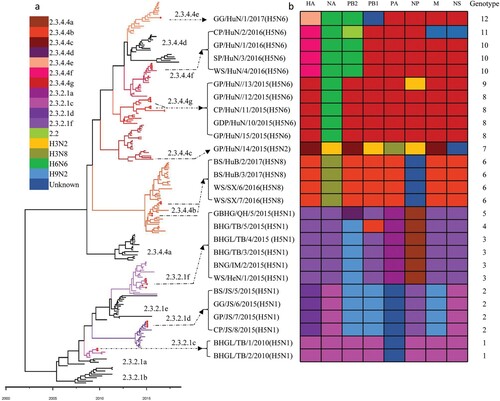
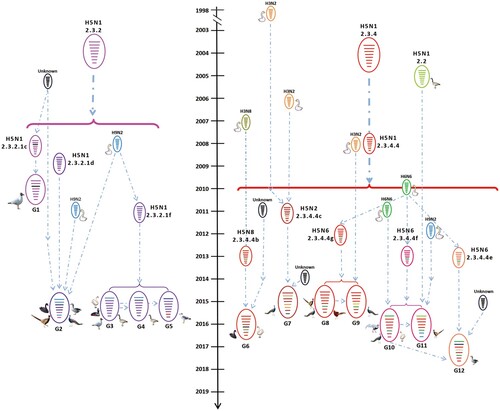
Figure 3. Diagrammatic representation of the formation of different H5 reassortants. Viral particles are represented by coloured ovals containing horizontal bars representing the 8 gene segments (from top to bottom: PB2, PB1, PA, HA, NP, NA, M, and NS). Segments in descendant viruses are coloured according to their corresponding source virus (top) to illustrate gene ancestry through reassortment events. Possible donor viruses are adjacent to arrow tails; arrowheads point to the resulting reassortants. The timeline in the middle indicates the possible time of virus emergence or reassortment events. The tMRCA was estimated by using the Bayesian Markov chain Monte Carlo method in the BEAST v1.8.4 software package.