Figures & data
Table 1. Minimum inhibitory concentrations (MICs) or minimum effective concentrations (MECs) of different antifungal agents against Pleurostoma species.Table Footnotea
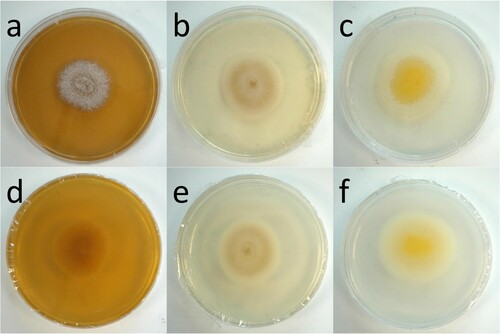
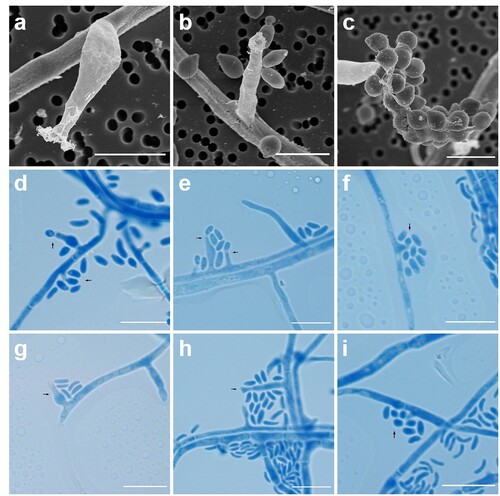
Table 2. Microscopic features of different Pleurostoma species during asexual life cycles.
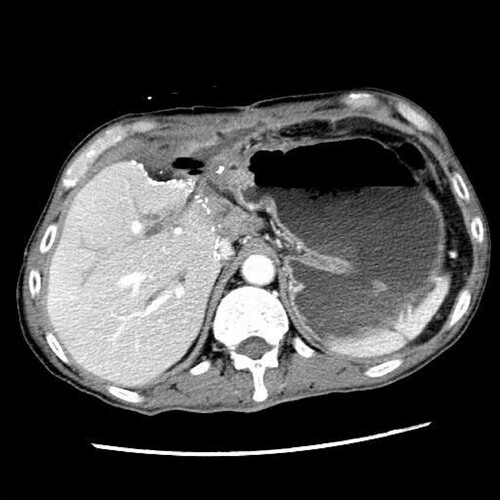
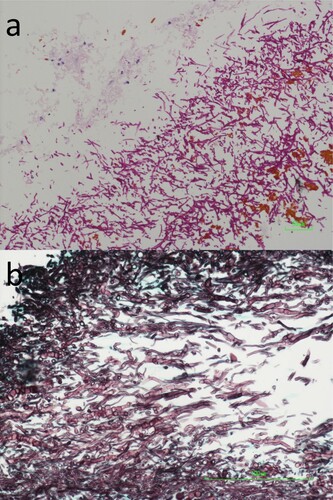
Table 3. Infections caused by Pleurostoma species.