Figures & data
Table 1. Biological properties of H9N2 viral NA-specific mAbs generated in this study.
Figure 1. Phylogenetic analysis of NA genes of 19 H9N2 field strains isolated from 1999 to 2019. Green dots represent vaccine strains. Red dot represents XXM H9N2 virus and black dots represent the other 18 field strains used in this study. The phylogenetic tree was constructed with MEGA X in neighbour-joining method and 1000 boot-strap replicates.
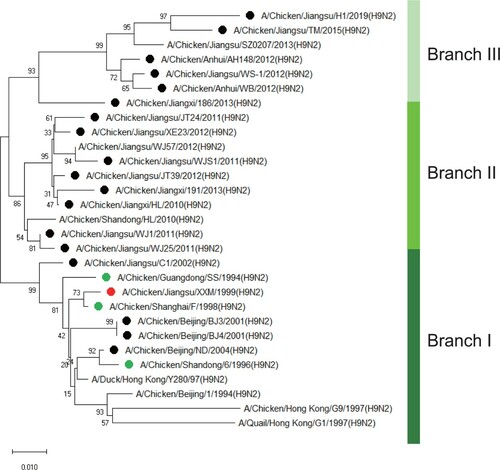
Table 2. Reactivity of 22 N2-specific mAbs to 19 field strains of H9N2 isolated from 1999 to 2019.
Figure 2. The impact of N356D mutation on reactivity of group I mAbs. (A) Amino acid sequence alignment of NA sequences (342–379, N2 numbering) of 19 field strains was analysed with MEGA X. The D356 in NA of A/Chicken/Jiangsu/C1/2002 (H9N2) virus was marked in dark blue. (B) Plasmids of WT NA gene (pCAGGS-NA (N356)) and mutant NA gene (pCAGGS-NA (D356)) were transfected into COS-1 cells and examined with 4 mAbs in IFA.
Table 3. Natural substitutions at 9 key amino acid positions of H9N2 AIV field strains isolated in China.
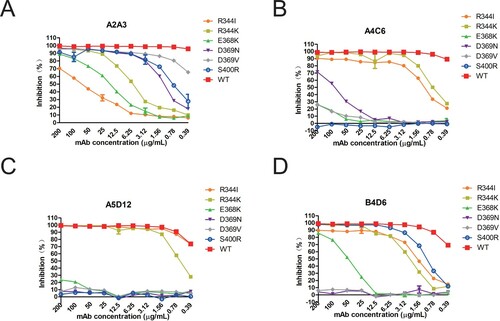
Figure 3. NI ability of group II mAbs to escape mutants and WT XXM H9N2 virus. NA inhibitory effect of mAbs A2A3, A4C6, A5D12 and B4D6 on escape mutants was measured in ELLA. A to D represent results of A2A3, A4C6, A5D12 and B4D6.