Figures & data
Figure 1. HBV core protein I126A mutation facilitates mature nucleocapsid uncoating.
Note: (A) HepG2 cells were infected with Ad-HBV or Ad-HBV I126A at an MOI of 10 and cultured in the absence or presence of 1μg/mL tetracycline (tet). HBV core DNA was extracted at day 6 post infection (dpi) and analysed by Southern blot hybridization. (B) HepG2 cells were infected with Ad-HBV or Ad-HBV I126A at an MOI of 10. The cells were treated with 2 mM PFA from day 2 to day 6 post infection. Cells were then harvested and cytoplasmic HBV capsids were purified by sucrose gradient centrifugation. After endogenous DNA polymerase reaction, capsid DNA were extracted with or without prior DNase I digestion and analysed by Southern blot hybridization. RC, relaxed circular DNA; DSL, double-stranded linear DNA; SS, single-stranded DNA.
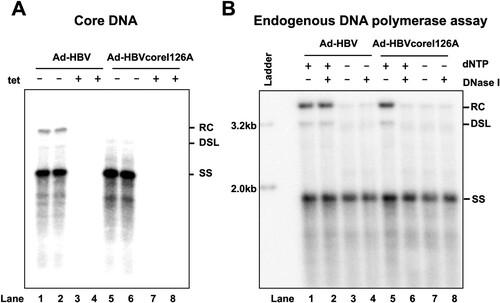
Figure 2. HBV core DNA can activate cGAS-STING pathway upon transfection into human hepatoma cells reconstituted with functional cGAS and STING.
Note: Parental HepG2, HepG2/cGAS-STING and HepG2/cGAS-STINGΔC cells were mock-transfected or transfected with 1 μg HBV core DNA or dsDNA-90 per well of 12-well plates and harvested at 8 h post transfection. The levels of IL-29 (A) and CXCL10 (B) mRNA were determined by qRT-PCR assays, normalized to β-actin mRNA and presented as the fold of induction over that in mock-transfected HepG2 cells. Mean and standard deviations are presented (n = 3).
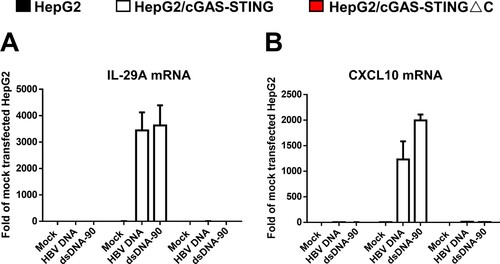
Figure 3. Uncoating of mature HBV nucleocapsids did not activate cGAS-STING pathway.
Note: Parental HepG2, HepG2/cGAS-STING, HepG2/cGAS-STING△C cells were infected with Ad-HBV or Ad-HBV I126A at an MOI of 10 and culture in the absence or presence of tet. At day 5 post infection, the cells were mock-treated or treated with 1 μM of GLS4 for 24 h. Cytoplasmic core DNA were extracted and analysed by Southern blot hybridization (Panels (A)–(C)). Total cellular RNA were extracted and the levels of HBV pgRNA (D), IL-29 (E) and CXCL10 (F) mRNA were determined by qRT-PCR assays, normalized to β-actin mRNA and presented as the fold of induction over that in mock-transfected HepG2 cells cultured in the presence of tet. Mean and standard deviations are presented (n = 3). Black, white and red bars denote parental HepG2, HepG2/cGAS-STING, and HepG2/cGAS-STING△C cells, respectively.
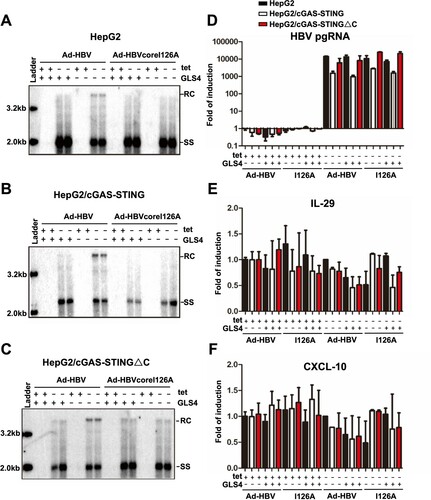
Figure 4. Mature nucleocapsid uncoating supports cccDNA biosynthesis in human hepatocytes.
Note: (A) HepG2 cells were infected with Ad-HBV or Ad-HBV-I126A at an MOI of 10 and cultured in the absence of tet. The cells were harvested at day 6 post infection and Hirt DNA was extracted and analysed by Southern blot assay without treatment (lanes 1 and 4), after denaturation at 88°C for 8 min (lanes 2 and 5) or EcoRI digestion after heat denaturing (lanes 3 and 6). (B and C) HepG2 and PXB cells were infected with Ad-HBV or Ad-HBV-I126A at an MOI of 10 and cultured in the absence of tet. The Cells were mock-treated or treated with 10 μM of lamivudine (HepG2) or 1 μM of entecavir (PXB cells) from day 2 to day 6 after infection. HBV core DNA and Hirt DNA were extracted and analysed by Southern blot hybridization. Hirt DNA extracted from the cells were denatured at 88°C for 8 min to denature DP-rcDNA to single-stranded DNA and followed by restriction with EcoRI to convert cccDNA into unit-length double DNA.
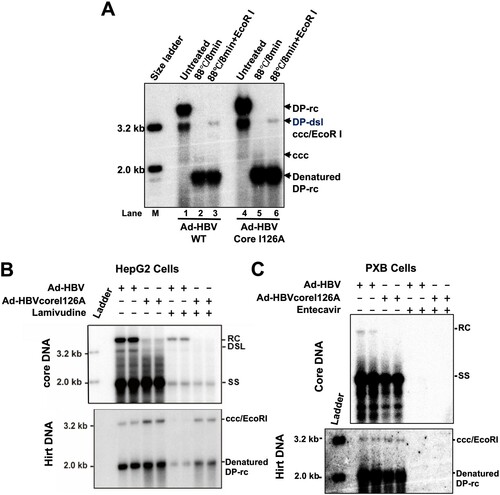
Figure 5. Over-expression of TREX1 selectively digests mature double-stranded HBV DNA in HepAD38 cells.
Note: (A) Expression of wild-type and mutant TREX1 in the indicated cell lines was detected by a western blot assay with an antibody against TREX1. β-actin served as a loading control. (B) Cytoplasmic localization of wild-type and mutant TREX1 in the indicated cell lines was indicated by an immunofluorescent assay with an antibody against TREX1. (C) Hirt DNA extracted from the indicated cells were denatured at 88°C for 8 min to denature DP-rcDNA to single-stranded DNA and followed by restriction with EcoRI to convert cccDNA into unit-length double stranded linear DNA and detected by Southern blot hybridization. Host cellular mitochondrial DNA served as loading controls for Hirt DNA.
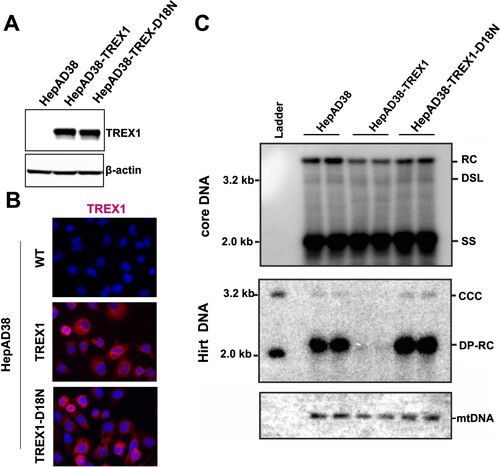
Figure 6. Endogenous TREX1 does not play a significant role in elimination of cytoplasmic double-stranded HBV DNA in GLS4-treated cells.
Note: Parental HepAD38 and its derived cell lines were cultured in the absence of tet for 6 days. The cells were then mock-treated or treated with 5 μM of GLS4 for 24 h. Cytoplasmic HBV core DNA were extracted from the cells and determined by Southern Blot hybridization.
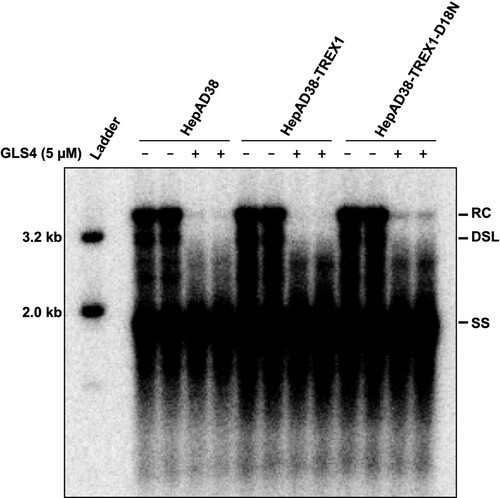
Figure 7. Expression of TREX1 does not inhibit the synthesis of cccDNA in de novo infection.
Note: (A) Expression of wild-type and mutant TREX1 in the indicated cell lines were detected by a western blot assay with an antibody against TREX1. β-actin served as a loading control. (B) Cytoplasmic localization of wild-type and mutant TREX1 in the indicated cell lines were indicated by an immunofluorescent assay with an antibody against TREX1. (C) Schematic presentation of HBV infection and treatment schedule. (D) Hirt DNA were extracted at 12 and 72 h post infection and analysed by Southern blot hybridization.
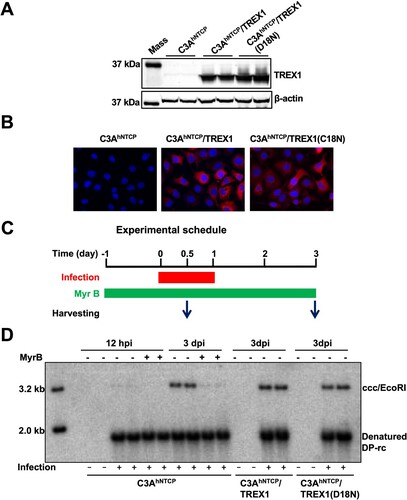
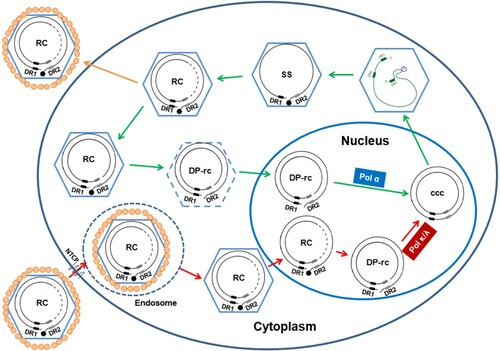
Figure 8. Schematic illustration of the difference in de novo cccDNA synthesis and intracellular cccDNA amplification pathways.
Note: The nucleocapsid with dashed line indicates partially disassembled nucleocapsid. The DP-rcDNA associated with the partially disassembled nucleocapsid is sensitive to nuclease digestion, but can be imported into the nucleus for cccDNA synthesis. DR1 and DR2 indicate the direct repeat DNA sequence 1 and 2, respectively. The black filled dot at the 5′ end of minus strand DNA represents viral DNA polymerase. See text in introduction section for detailed explanation of viral replication cycle.