Figures & data
Figure 1. Shared VH3-53-derived antibodies were observed in individuals exposed to SARS-CoV-2. (A) Scheme of longitudinal sample collection and antibody repertoire sequencing. Circles indicate samples used for both heavy- and light-chain repertoire sequencing, while triangles indicate samples used for only heavy-chain repertoire sequencing. (B) Comparison of VH3-53 germline gene usage in heavy chain repertoires between different time points after the onset of symptoms. Statistical tests were performed by Student’s t-test (Unpaired, two-tailed). (C) Scheme of V(D)J gene rearrangement and identification of shared clonotype. Shared clonotype are defined as a group of sequences with the same V/J combination event, the same heavy chain complementary determining region (HCDR3) length, and HCDR3 amino acid sequences of 80% identity between different people. (D) Lineage structure of the VH3-53-J6 clonotype. Each dot represents a unique antibody clone, two dots are connected via a line if they have 80% amino acid similarity on HCDR3 region, the dot area represents the read count for each unique antibody clone, and different colours were used to distinguish different COVID-19 donors and healthy controls. (E) The HCDR3 sequence comparison of the VH3-53-J6 clonotype in the repertoires of different COVID-19 donors. (F) Presence of the shared VH3-53-J6 clonotype in repertoires of COVID-19 patients and healthy people. (G) Identity-divergence plot of three VH3-53 antibody clones (PtL108-83, PtZ108-117, and PtK111-203) from samples collected three months after hospital discharge. All VH3-53 sequences in the repertoires were plotted as a function of sequence divergence from VH3-53 germline gene and sequence identity to PtL108-83, PtZ108-117, and PtK111-203, respectively. Colour gradient indicated sequence density. The sequence with an identical HCDR3 with PtL108-83, PtZ108-117, and PtK111-203 are shown as magenta dots.
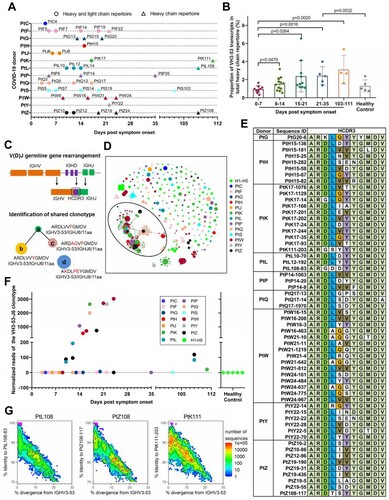
Figure 2. Sequence analysis of the VH3-53-J6 clonotype. Maximum-likelihood (ML) tree (left panel) and multiple alignments of amino acid sequences (middle panel) are shown. Somatic hypermutation rates of each sequence are also shown (right panel). Different coloured circles in the ML tree represent sequences that derived from different COVID-19 patients. Different coloured lines represent sequences that belong to different clades.
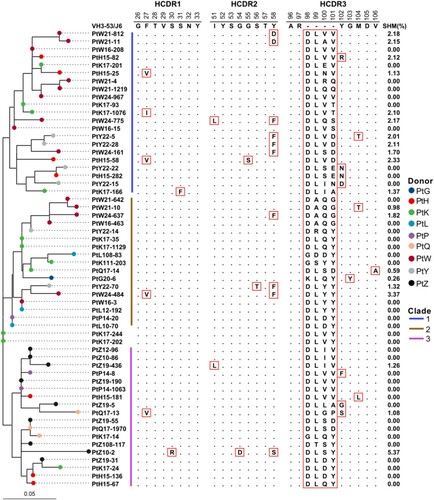
Figure 3. Scheme of germline gene rearrangements. Inferred germline V(D)J genes of closely related the VH3-53-J6 clonotype identified from different patient’s repertoires. Each block is labelled with the inferred germline sequences. Nucleotide positions and corresponding amino acid positions are labelled in blue colour if the sequences are same as the germline genes (top of each block). Nucleotide positions and corresponding amino acid positions are not coloured if they are inferred to be derived from un-templated nucleotide addition during V(D)J combination. Nucleotide positions are coloured with red colour if one amino acid is encoded by different synonymous codons. (A) Comparison of V(D)J gene rearrangement of PtH15-136, PtK17-24, PtL10-70, PtP14-20, and PtZ19-31. (B) Comparison of V(D)J gene rearrangement of PtL12-192, PtW24-484, and PtY22-70. (C) Comparison of V(D)J gene rearrangement of PtP14-1063, PtW16-208, PtW24-967, and PtZ19-190. (D) Comparison of V(D)J gene rearrangement of PtQ17-1970, PtW21-161, and PtZ19-55.
Figure 4. Light-chain VK1-9-encoded antibodies were highly shared among COVID-19 patients. (A) The usage distribution of light-chain germline gene that paired with VH3-53 in 80 known RBD-targeted antibodies[Citation3, Citation4, Citation5, Citation6, Citation7, Citation8, Citation9, Citation10, Citation11, Citation12, Citation13, Citation14]. (B) Comparison of LCDR3 sequences of the known VK1-9-encoded light chains and their presence in the repertoires of COVID-19 patients and healthy people. (C) Comparison of LCDR3 sequences of the shared VK1-9-J4 clonotype identified from the repertoires of COVID-19 patients and healthy people. (D) The proportion of the VK1-9-J4 clonotype in the total VK1-9 light-chain repertoires of COVID-19 patients and healthy people.
![Figure 4. Light-chain VK1-9-encoded antibodies were highly shared among COVID-19 patients. (A) The usage distribution of light-chain germline gene that paired with VH3-53 in 80 known RBD-targeted antibodies[Citation3, Citation4, Citation5, Citation6, Citation7, Citation8, Citation9, Citation10, Citation11, Citation12, Citation13, Citation14]. (B) Comparison of LCDR3 sequences of the known VK1-9-encoded light chains and their presence in the repertoires of COVID-19 patients and healthy people. (C) Comparison of LCDR3 sequences of the shared VK1-9-J4 clonotype identified from the repertoires of COVID-19 patients and healthy people. (D) The proportion of the VK1-9-J4 clonotype in the total VK1-9 light-chain repertoires of COVID-19 patients and healthy people.](/cms/asset/614613c6-56d2-4d15-aa8d-8635f21f14d6/temi_a_1925594_f0004_oc.jpg)
Figure 5. VH3-53-J6 antibodies identified from the repertoires of different COVID-19 patients can bind to SARS-CoV-2 RBD and block the interaction between RBD and ACE2. (A) Eight recombinant VH3-53 antibodies with relatively higher RBD-binding activities were selected for further analysis. Previously published mAbs B-38 [Citation11], CR3022 [Citation33] and 6A6 [Citation47] were used as controls. (B) Binding kinetics of the 8 recombinant VH3-53 antibodies with SARS-CoV-2 RBD were measured by BLI. Immobilized individual antibody (10 μg/mL) was saturated with RBD at seven different concentrations (200 nM, 100 nM, 50 nM, 25 nM, 12.5 nM, 6.25 nM, and 3.125 nM) or buffer. (C) Neutralizing activities of the 8 recombinant VH3-53 antibodies, determined using a SARS-CoV-2 surrogate virus neutralization test (sVNT). B-38, CR3022 and 6A6 were used as controls (D) Comparison of B-38 (PDB: 7BZ5), CV30 (PDB: 6XE1) [Citation17], CC12.3 (PDB: 6XC4) [Citation16], CR3022 (PDB: 6W41), and P2B-2F6 (PDB: 7BWJ) [Citation3] binding epitopes. SARS-CoV-2 RBD is shown in cartoon representation (grey). The heavy and light chains of B-38, CV30, CC12.3, CR3022, and P2B-2F6 are coloured in cyan, limegreen, skyblue, magenta, and yellow, respectively. (E) Crystal structure of ACE2 (green) in complex with SARS-CoV-2 RBD (grey) (PDB: 6LZG) [Citation1]. (F) Comparison of rmAb23, rmAb33, rmAb12, B-38, CV30, and CC12.3 binding epitopes. Modelling structure of rmAb23, rmAb33, and rmAb12 in complex with SARS-CoV-2 RBD are coloured in orange, wheat, yellow-orange and grey, respectively. (G) Competition binding to RBD between the 8 recombinant VH3-53 antibodies and B-38, CR3022, or 6A6 was measured by BLI, respectively. Immobilized RBD (2 μg/mL) was saturated with 10 μg/mL of the first antibody and then flowed with equal concentration of the first antibody in the presence of or without the second antibody. The graphs show binding patterns after saturation of RBD.
![Figure 5. VH3-53-J6 antibodies identified from the repertoires of different COVID-19 patients can bind to SARS-CoV-2 RBD and block the interaction between RBD and ACE2. (A) Eight recombinant VH3-53 antibodies with relatively higher RBD-binding activities were selected for further analysis. Previously published mAbs B-38 [Citation11], CR3022 [Citation33] and 6A6 [Citation47] were used as controls. (B) Binding kinetics of the 8 recombinant VH3-53 antibodies with SARS-CoV-2 RBD were measured by BLI. Immobilized individual antibody (10 μg/mL) was saturated with RBD at seven different concentrations (200 nM, 100 nM, 50 nM, 25 nM, 12.5 nM, 6.25 nM, and 3.125 nM) or buffer. (C) Neutralizing activities of the 8 recombinant VH3-53 antibodies, determined using a SARS-CoV-2 surrogate virus neutralization test (sVNT). B-38, CR3022 and 6A6 were used as controls (D) Comparison of B-38 (PDB: 7BZ5), CV30 (PDB: 6XE1) [Citation17], CC12.3 (PDB: 6XC4) [Citation16], CR3022 (PDB: 6W41), and P2B-2F6 (PDB: 7BWJ) [Citation3] binding epitopes. SARS-CoV-2 RBD is shown in cartoon representation (grey). The heavy and light chains of B-38, CV30, CC12.3, CR3022, and P2B-2F6 are coloured in cyan, limegreen, skyblue, magenta, and yellow, respectively. (E) Crystal structure of ACE2 (green) in complex with SARS-CoV-2 RBD (grey) (PDB: 6LZG) [Citation1]. (F) Comparison of rmAb23, rmAb33, rmAb12, B-38, CV30, and CC12.3 binding epitopes. Modelling structure of rmAb23, rmAb33, and rmAb12 in complex with SARS-CoV-2 RBD are coloured in orange, wheat, yellow-orange and grey, respectively. (G) Competition binding to RBD between the 8 recombinant VH3-53 antibodies and B-38, CR3022, or 6A6 was measured by BLI, respectively. Immobilized RBD (2 μg/mL) was saturated with 10 μg/mL of the first antibody and then flowed with equal concentration of the first antibody in the presence of or without the second antibody. The graphs show binding patterns after saturation of RBD.](/cms/asset/3acf014e-043c-476d-8743-1b9adfaee8f3/temi_a_1925594_f0005_oc.jpg)
Table_S1_editable.xlsx
Download MS Excel (14.4 KB)Data availability statement
All materials that supported this study are available from the corresponding authors upon reasonable request. All raw antibody repertoire data used in this study have been deposited at the National Genomics Data Center (https://bigd.big.ac.cn/) under the accession number: PRJCA003775.
