Figures & data
Figure 1. Immunization strategy of DNA prime followed by protein boost successfully induced SARS-CoV-2 specific and neutralizing antibody responses. (A) Two rabbits received three doses of DNA vaccine encoding the SARS-CoV-2 RBD on day 0, day 7 and day 21, and were further boosted with two doses of the SARS-CoV-2 S1 protein emulsified with incomplete Freund’s adjuvant (IFA) on day 35 and day 49. Sera were collected on day 0, day 28, and day 56 for immunoassays. (B) The serological IgG titer specific to RBD, S1 and ECD protein from naive rabbits (day 0) and immunized rabbits (days 28 and 56).
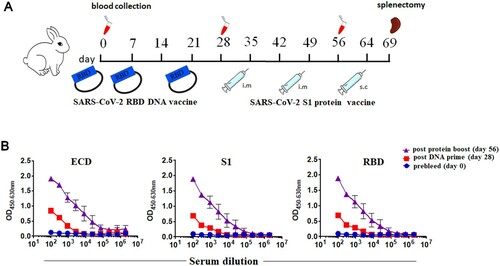
Figure 2. Functional screening of cultured S1-specific single rabbit B cell clones. (A) The sorting strategy for S1-specific B cells from rabbit splenocytes using FACS. (B) High-throughput functional screening of cultured B cells. The number of individually cultured B cell clones (Cultured B cell clones), B cell clones binding SARS-CoV-2 S1 protein (SARS-CoV-2 S1 binding clones), B cell clones binding the SARS-CoV-2 RBD (SARS-CoV-2 RBD binding clones), B cell clones with neutralization potential (Neutralization clones), and B cell clones with the capability of inhibiting RBD and ACE2 interaction (RBD/ACE2 blocking clones) were listed. (C) Representative ELISA screening of cultural supernatants for top 40 B cell clones that can bind to the SARS-CoV-2 S1 protein. (D) Pseudovirus neutralization assay of S1-specific B cell cultural supernatants. (E) ACE2 blocking assay of S1-specific B cell cultural supernatants.
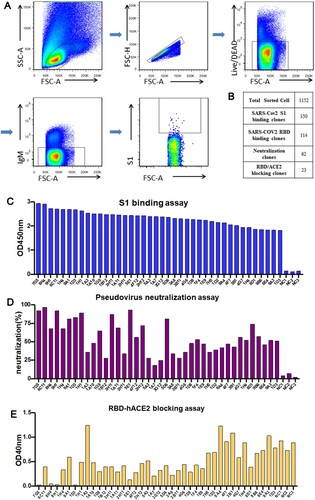
Figure 3. Binding of four neutralizing RmAbs, 1H1, 9H1, 7G5, and 5E1 to SARS-CoV-2 spike proteins and RBD mutants. (A) Four clones of RmAbs bind to purified SARS-CoV-2 ECD, S1 and RBD in ELISA. (B) RmAbs have different recognition of RBD and RBD mutants including RBD N501Y, RBD K417N, RBD E484K, and RBD N501Y/K417N/E481K. (C) RmAbs bind to S1 and S1 mutants S1 69-70 deletion. (D) RmAbs differentially recognize the SARS-CoV-2 RBD protein in Western Blotting.
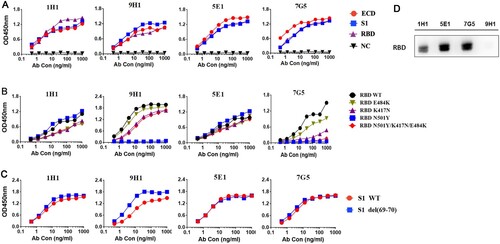
Figure 4. Binding kinetics of RmAbs to SARS-CoV-2 S1 and RBD proteins. (A) Surface plasmon resonance sensorgrams demonstrate the binding and dissociation kinetics of RmAbs to SARS-CoV-2 S1 and RBD proteins. Changes in resonance units (RU) over time are indicative of binding kinetics. (B) Rate constant Ka (equilibrium association constant), Kd (dissociation constant) and KD of RmAbs were determined respectively when binding to the SARS-CoV-2 S1 and RBD.
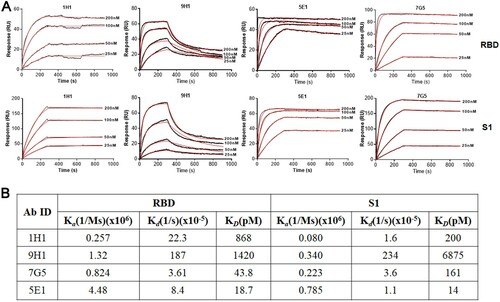
Figure 5. The neutralization potency of four RmAbs against SARS-CoV-2 pseudovirus variants and live viruses. (A) SARS-CoV-2 pseudovirus neutralization assay of four RmAbs (9H1, 1H1, 5E1 and 7G5) against wild type (WT) strain, the D614G variant, the B.1.1.7 variant, the B.1.429 variant, the P.1 variant, the B.1.526 variant, and the B.1.351 variant. The curve is presented by the inhibition percentage of SARS-CoV-2 pseudotyped viruses entry into host cells. (B) Neutralizing potency (IC50) of RmAbs was determined using either seven SARS-CoV-2 pseudotyped variants or SARS-CoV-2 live viruses.
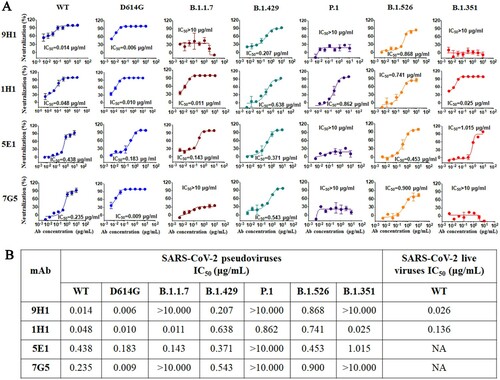
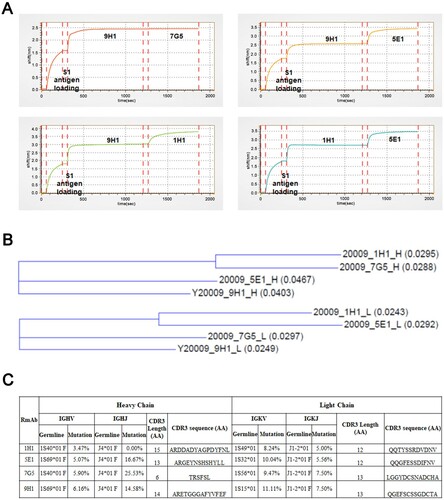
Figure 6. Epitope binning and sequence analysis of neutralizing RmAbs. (A) Epitope binning assay was performed to assess the pairing of mAbs when binding to the SARS-CoV-2 S1 protein. Binding intensity was indicated as the shift in nm over time. (B) Phylogenetic analysis of heavy chain (above) and light chain (below) genes for four neutralizing RmAbs using the maximum-likelihood method. Branch lengths are drawn to scale to visualize sequence diversification. (C) Mutational status of the variable regions, joining regions, and CDR3 regions of RmAb heavy chains and light chains compared to matched germline clone sequences. IGHV: Immunoglobulin heavy-chain variable region, IGHJ: Immunoglobulin heavy-chain joining region; IGKV: Immunoglobulin kappa chain variable region; IGKJ: Immunoglobulin kappa chain joining region.