Figures & data
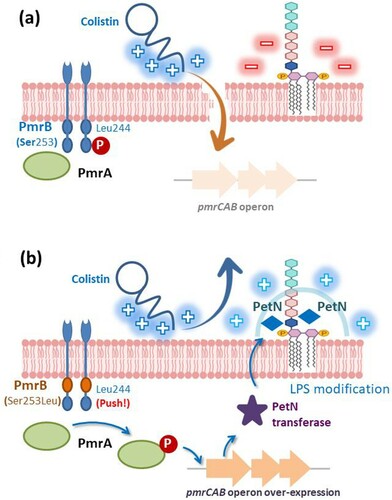
Figure 1. Relative expression of pmrC, pmrA, lpxC, and lpxA genes in Acinetobacter nosocomialis isolates. Each isolate was tested in triplicate in two independent experiments. Bars represent the average, and error bars represent standard deviations. Black bars, colistin-resistant Acinetobacter nosocomialis (ColRAN) isolates; white bars, colistin-susceptible Acinetobacter nosocomialis (ColSAN) isolates. Data were analysed using an independent t-test (*p <0.05).
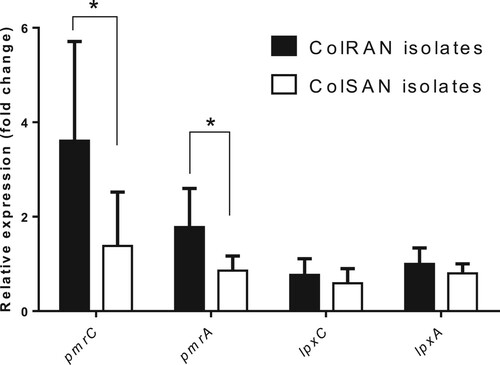
Table 1. Pasteur multi-locus sequence typing profile and amino acid variation patterns in PmrA and PmrB proteins.
Table 2. Relationship between colistin minimum inhibitory concentrations and expression of pmrC, pmrA, lpxC, and lpxA genes in various transformants.
Figure 2. Structural models of PmrB. (a) The modelling structure of the Type 1 PmrB homodimer is shown as a cartoon diagram; one subunit is shown in light pink and the other is shown in light yellow. (b, c, and d) Close-up view of two types of orientations of Ser253Leu (Type 2a and Type 2b) with substitution of PmrB. The side chains of His236 and His236' are shown as green and cyan stick models, respectively. The carbon atoms of Ile243, Leu244, Ser253, and L253 are shown as blue ball and stick models, respectively. The carbon atoms of Ile243', Leu244', Ser253', and L253' are shown as cyan ball and stick models. Oxygen atoms are shown in red. The steric collisions between L253 and L244' or L253' and L244 of Type 2b are indicated by arrows (b).
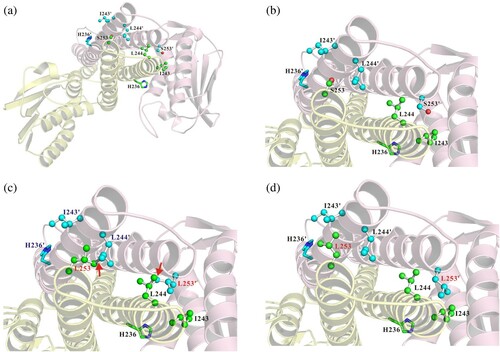
Figure 3. Schematic representation for the mechanism of colistin resistance. (a) Colistin is a cationic antimicrobial peptide. Colistin targets the anionic lipid A portion of lipopolysaccharides (LPS) and binds to phospholipids in bacterial cell membranes. This binding leads to changes in the permeability of the outer cell membrane and leakage of cell contents. The PmrAB locus is a two-component system (TCS) that can regulate the expression of the pmrCAB operon. The pmrCAB operon usually has a low expression level and encodes three functional proteins, including PmrC (PetN transferase), PmrA (response regulator), and PmrB (sensor kinase). (b) A model based on our data: amino acid substitution in PmrB (Ser253Leu) caused overexpression of the pmrCAB operon. Overexpression of the pmrC gene will generate several phosphoethanolamine (PetN) transferases. PetN transferase can add PetN to either the 4' or 1' phosphate of lipid A in LPS. This modification of LPS results in positively charged phosphate groups and prevents the binding of the cationic colistin.