Figures & data
Figure 1. V. vulnificus phylogeny. The phylogenetic tree was reconstructed using the maximum-likelihood method and the generalized time-reversible model (GTR + F+R5) of evolution. Bootstrap support values from 1000 replicates are indicated in the corresponding nodes as percentages. L, lineage.
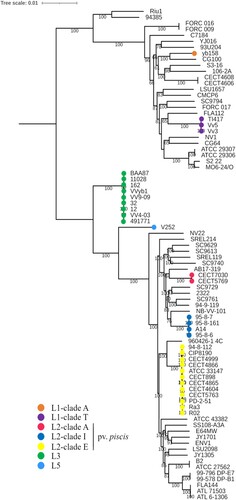
Figure 2. Plasmids in pv. piscis lineages and clades. The genes for conjugative transference, the survival in fish blood kit (ftpb and fpcrp), and the MARTX toxin are represented in green, red, and orange respectively, while the additional genes common to all pv. piscis plasmids are represented in brown. (A) Linear comparison among pCladeT and the original pv. piscis plasmids, pFv and pConj, performed with Easyfig [Citation38]. (B) Ring representation of the pv. piscis plasmids from the clades and lineages emerged in the Eastern Mediterranean. From inside to outside, pL3 (used as reference; black ring), pClade A, pCladeT y pL5. The gene annotation of the pL3 is represented in the multicoloured external ring.
![Figure 2. Plasmids in pv. piscis lineages and clades. The genes for conjugative transference, the survival in fish blood kit (ftpb and fpcrp), and the MARTX toxin are represented in green, red, and orange respectively, while the additional genes common to all pv. piscis plasmids are represented in brown. (A) Linear comparison among pCladeT and the original pv. piscis plasmids, pFv and pConj, performed with Easyfig [Citation38]. (B) Ring representation of the pv. piscis plasmids from the clades and lineages emerged in the Eastern Mediterranean. From inside to outside, pL3 (used as reference; black ring), pClade A, pCladeT y pL5. The gene annotation of the pL3 is represented in the multicoloured external ring.](/cms/asset/33730334-60c1-4510-9755-10f105d33571/temi_a_1999177_f0002_oc.jpg)
Table 1. Isolation data, lineage and identification of the new isolates in comparison with those of the control strains.
Table 2. Results obtained in the in vivo and ex vivo virulence assays performed with the new isolates in comparison with those of the control strains.
Figure 3. rtxA1 gene structure. (A) Schematic representation of rtxA1 gene from pv. piscis L2-clade E and L1-clade T (rtxA1 L2-clade E and rtxA1L1-clade T) (B) Comparison among the rtxA1 genes of the clades and lineages emerged in the Eastern Mediterranean. The images were constructed with Easyfig and the blue scale indicates nucleotide Blast similarity. Carboxi and amino terminal modules are coloured in grey, cysteine protease domain (CPD) in turquoise, Domain X (DmX) in pink, ExoY-like adenylate cyclase domain (ExoY) in black, makes caterpillars floppy-like domain (MCF) in orange, alpha/beta hydrolase domain (ABH) in purple, Rho GTPase-inactivation domain (RID) in green and domain of unknown function at the first position (DUF1) in yellow.
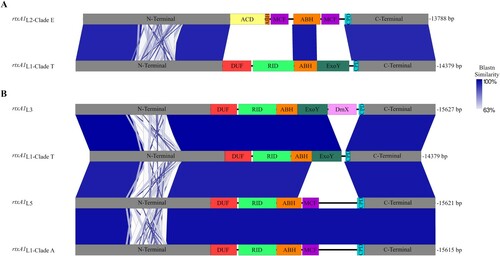
Table 3. Genotypic and phenotypic characterization of selected strains from previously described lineages and/or clades possessing a pFv-related plasmid.
Table 4. In vivo and ex vivo virulence assays performed with selected strains from previously described lineages and/or clades possessing a pFv-related plasmid.
Figure 4. Evolutionary history of the genes for the “survival in fish blood kit”. Molecular phylogenetic analysis of ftbp (A) and fpcrp (B) was performed using the maximum likelihood method based on the Tamura 3-parameter model [Citation47]. The tree is drawn to scale, with branch lengths measured as the number of substitutions per site. The main host (tilapia or eel) is shown in each tree.
![Figure 4. Evolutionary history of the genes for the “survival in fish blood kit”. Molecular phylogenetic analysis of ftbp (A) and fpcrp (B) was performed using the maximum likelihood method based on the Tamura 3-parameter model [Citation47]. The tree is drawn to scale, with branch lengths measured as the number of substitutions per site. The main host (tilapia or eel) is shown in each tree.](/cms/asset/04307061-04aa-4d5f-842f-f9f827b545ee/temi_a_1999177_f0004_oc.jpg)
