Figures & data
Figure 1. Schematic illustration of the spike protein of the SARS-CoV-2 Lambda variant (A) Domain structure of the Lambda variant, including amino acid changes. Circles indicate an amino acid mutation at that position; a mutation with seven amino acid deletions is indicated by underlined text. NTD, N-terminal domain; RBD, receptor-binding domain; FP, fusion peptide; HR1, heptad repeat 1; HR2, heptad repeat 2; CP, cytoplasmic domain. The S1/S2 cleavage site is indicated with a double slash at the 685/686 site of the Spike protein. (B) The 3D structure of the Lambda variant.
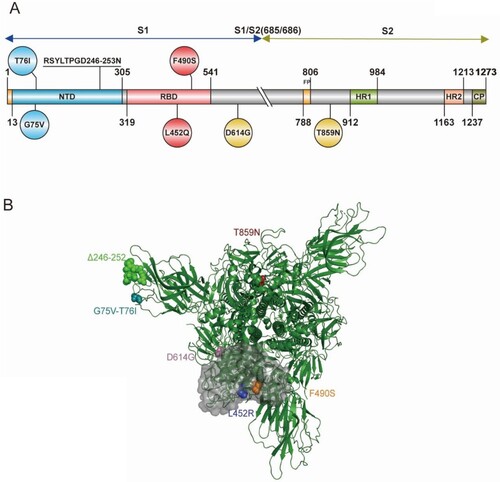
Figure 2. Infectivity analysis of Lambda variant pseudoviruses Infection assays with the 10 Lambda variant-related mutant pseudotyped viruses in Huh-7 (A), Calu-3 (B), Vero (C), LLC-MK2 (D), 293T-ACE2 (E), 293T-ACE2-Furin (F), 293T-ACE2-TMPRSS2 (G) and 293T-ACE2-CatL (H) cells. The relative infectivity of the mutant pseudoviruses is presented as the ratio of relative luciferase units (RLU) for mutant/RLU for the D614G variant. All results were obtained from three independent experiments (mean ± SEM). The dashed lines indicate the threshold of four-fold differences. One-way ANOVA and Dunnett's multiple comparisons test were employed to determine differences in infectivity between Lambda and D614G.
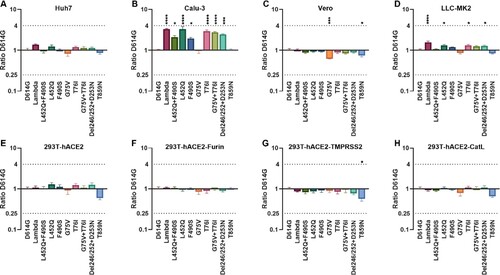
Figure 3. Analyses of Lambda variant antigenicity using a panel of neutralizing mAbs (A) Heatmap representation of neutralization reactions using 12 neutralizing mAbs against 10 Lambda-related mutant pseudoviruses. The ratio of ID50 value between Lambda and the D614G reference was calculated and analysed, followed by heatmap construction using HemI software [Citation38]. Data represent three independent experiments. Red and blue boxes represent increased and decreased viral sensitivity to mAbs, respectively, as shown in the scale bar. A 4-fold difference was considered significant. Structural modelling of the L452Q and F490S mutations, based on RBD-9G11 for 9G11 (B), and RBD-X593 for X593 (C). The colour of the RBD ranges from yellow to white to blue indicating hydrophobic to hydrophilic, and the images were produced in UCSF ChimeraX.
![Figure 3. Analyses of Lambda variant antigenicity using a panel of neutralizing mAbs (A) Heatmap representation of neutralization reactions using 12 neutralizing mAbs against 10 Lambda-related mutant pseudoviruses. The ratio of ID50 value between Lambda and the D614G reference was calculated and analysed, followed by heatmap construction using HemI software [Citation38]. Data represent three independent experiments. Red and blue boxes represent increased and decreased viral sensitivity to mAbs, respectively, as shown in the scale bar. A 4-fold difference was considered significant. Structural modelling of the L452Q and F490S mutations, based on RBD-9G11 for 9G11 (B), and RBD-X593 for X593 (C). The colour of the RBD ranges from yellow to white to blue indicating hydrophobic to hydrophilic, and the images were produced in UCSF ChimeraX.](/cms/asset/215c80c2-c9a3-4ab9-8fd5-7cf85d757f52/temi_a_2008775_f0003_oc.jpg)
Figure 4. Analysis of the antigenicity of the Lambda variant using a panel of convalescent sera (A) Heatmap representation of the neutralization reactions using 14 convalescent sera (CS1-14) against 10 Lambda-related mutant pseudoviruses. The ratio of ID50 values (mean of three independent experiments) between the Lambda variant and D614G reference was calculated and analysed, followed by construction of a heatmap using HemI software. Red and blue boxes represent increased and decreased viral sensitivity to convalescent sera, respectively. (B) Violin plot of the summary and statistical analysis of changes in antigenicity of the Lambda variant. Each dot (mean of three independent experiments) represents one convalescent serum. One-way ANOVA and Holm-Sidak's multiple comparison tests were used to analyse the differences between groups. A P value less than 0.05 was considered statistically significant. * P<0.05, ** P<0.01, *** P<0.001, and **** P<0.0001. (C) Analysis of the ID50 values of neutralization reactions between the D614G reference and Lambda pseudoviruses to convalescent sera.
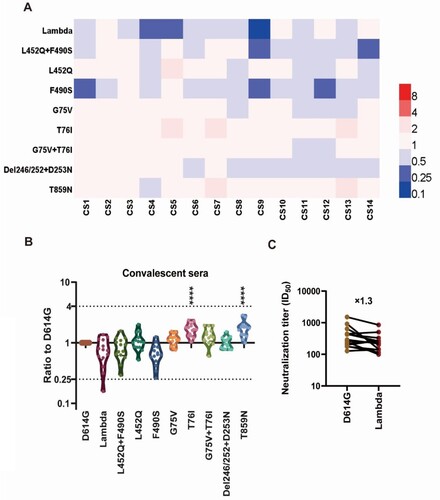
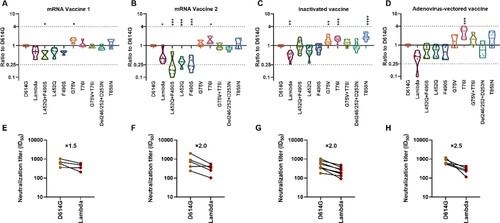
Figure 5. Analysis of the antigenicity of the Lambda variant using a panel of sera after immunization with different vaccine types Violin plot of the neutralization reactions against 10 Lambda-related mutant pseudoviruses using immune sera following immunization with different vaccine types (A, B, C, D). (A) mRNA vaccine 1: LPP/mRNA-Spike mRNA vaccine-immunized sera; (B) mRNA vaccine 2: ARCoV-RBD mRNA vaccine-immunized sera; (C) Inactivated vaccine: KCONVAC inactivated SARS-CoV-2 vaccine-immunized sera; (D) Adenovirus-vectored vaccine: Ad5 adenovirus-vectored vaccine-immunized sera. Analysis of the ID50 values of the neutralization reactions for vaccine-immunized sera between D614G and Lambda pseudoviruses (E, F, G, H). (E) mRNA vaccine 1: LPP/mRNA-Spike mRNA vaccine-immunized sera; (F) mRNA vaccine 2: ARCoV-RBD mRNA vaccine-immunized sera; (G) Inactivated vaccine: KCONVAC inactivated SARS-CoV-2 vaccine-immunized sera; (H)Adenovirus-vectored vaccine: Ad5 adenovirus-vectored vaccine-immunized sera. The data were the results from three replicates.