Figures & data
Figure 1. 2–36 neutralizes both SARS-CoV-2 and SARS-CoV. (A) Screening of mAb transfection supernatant for neutralizing activities against SARS-CoV-2 and SARS-CoV pseudoviruses. (B) 2–36 neutralization IC50 (µg/mL) against SARS-CoV-2 and SARS-CoV pseudoviruses (PV) as well as the live viruses (LV).
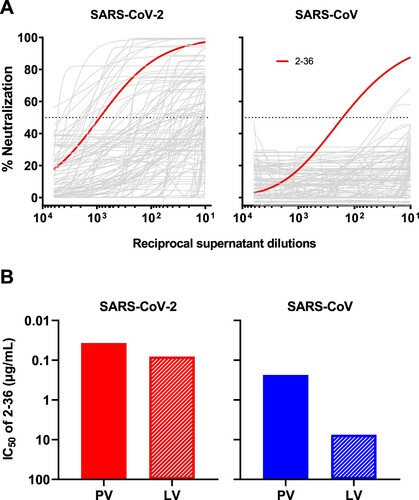
Figure 2. Cryo-EM structure of 2–36 in complex with SARS-CoV-2 and SARS-CoV Spike. (A) Cryo-EM reconstruction of 2–36 Fab in complex with the SARS-CoV-2 S trimer at 3.4 Å. RBD is coloured in green, the 2–36 Fab heavy chain in darker blue, the light chain in lighter blue, and the rest of the spike is coloured in light purple with glycans in darker purple. (B) Cryo-EM reconstructions of 2–36 Fab in complex with the SARS-CoV S trimer reveal two major classes: one 2–36 Fab bound to spike with 1-RBD up, and two 2–36 Fabs bound to spike with 2-RBD up. Reconstructions are shown in two different orientations. (C) The 2–36 interface model superposed onto an ACE2-RBD complex model (pdb: 6M0J) shows an ACE2 clash with the light chain. (D) Conservation analysis on the RBD among SARS-CoV-2, SARS-CoV, and bat and pangolin sarbecoviruses show 2–36 binding site is highly conserved, with regions of high conservation in grey and low conservation in red. The 2–36 epitope is outlined in dark blue, the epitope for COVA1-16 in cyan, and the interface with ACE2 in orange. (E) The interface model depicted in ribbon representation, with the CDR1 loops in gold, CDR2 loops in orange, and CDR3 loops in red. The interface residue contacts are shown for CDR H1, H3, and L2 complemented with the electron density map for a 4.1 Å local reconstruction. Hydrogen bonds between CDR H3 and RBD are depicted as dashed yellow lines.
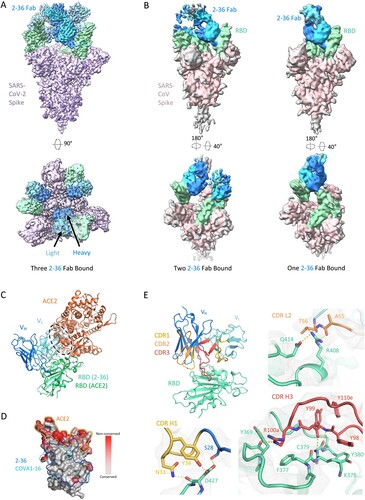
Figure 3. 2–36 neutralizes SARS-CoV-2 variants and SARS-CoV-like coronaviruses. (A) 2–36 neutralization IC50 (µg/mL) against SARS-CoV-2 variants. (B) Phylogenetic tree of SARS-CoV-2- and SARS-CoV-related lineages and other coronaviruses constructed via MEGA7 and maximum likelihood analysis of spike amino acid sequences extracted from the NCBI and GISAID database. Representative viruses selected for further testing are denoted in colour same as in (C). (C) Heatmap showing the neutralization IC50 values of the indicated antibodies against SARS-CoV-2, SARS-CoV and their related sarbecoviruses.
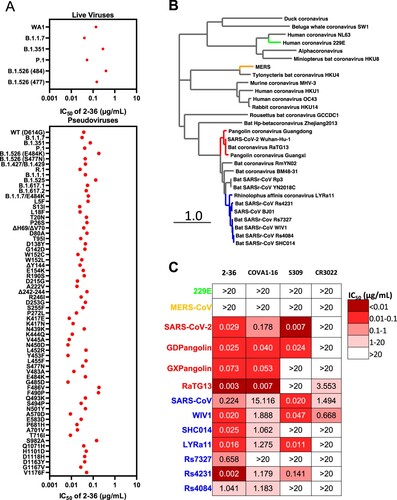
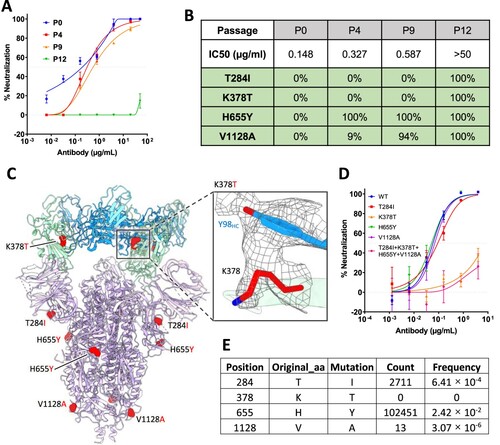
Figure 4. In vitro selection of 2–36 escape viruses. (A) Neutralizing activity of 2–36 against viruses at different passages. (B) Spike mutations found in viruses at different passages (C) Model of 2–36 in complex with the SARS-CoV-2 S trimer highlighting mutations in red. CDR H3 Tyrosine 98 makes van der Waals contacts with RBD residue 378 as shown as electron density map mesh in the subpanel. (D) The selected mutations were introduced into pseudoviruses and then tested for neutralization sensitivity to 2-36. (E) The frequency of the selected mutations in circulating in infected patients (data updated to Oct 13th, 2021).
Supplemental Material
Download MS Word (3.1 MB)Data availability
The cryo-EM structure of antibody 2–36 in complex with prefusion SARS-CoV-2 spike glycoprotein has been deposited in the PDB ID: 7N5H and EMDB ID: 24190.
