Figures & data
Figure 1. The comparison of the Mtb ribosome-bound clarithromycin (CTY) with the antibiotics from different ribosome complexes. (A) Chemical structures of NPET binding Erythromycin (ERY) and Clarithromycin; (B) The density map (deep blue mesh) of CTY in complex with the Mtb 50S ribosome viewed from two perspectives (maps contoured at 1.7σ and 1.2σ, respectively); (C) Comparison of the ribosome structures in complex with ERY or CTY (CTY carbon in yellow, PDB entry 7F0D, 50S of Mtb), ERY (carbon in green, PDB entry 4V7U, 70S of E. coli; carbon in cyan, PDB entry 6XHX, 70S of T. thermophilus) (D) Comparison of the 50S ribosome structures in complex with ERY or CTY (carbon in yellow, PDB entry 7F0D, Mtb), CTY (carbon in light pink, PDB entry 1J5A, D radiodurans) and ERY (carbon in grey, PDB entry 1JZY, D radiodurans).
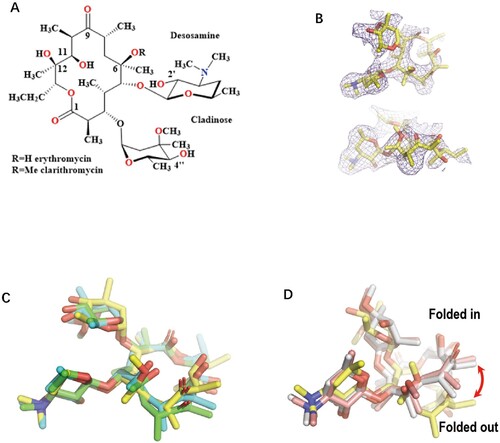
Figure 2. Structure of CTY in complex with the Mtb 50S ribosome and the close-up view. (A) Overview of the CTY binding site (red in square) in the Mtb 50S ribosome, view from the 30S side as indicated by the inset; Close-up views of CTY bound within the NEPT, together with two alternative conformations of A2062, non-swayed conformation in purple carbon and the swayed conformation of A2062 in green carbon (B) and (C); (D) The revised view with the surface (in cyan) for the macrolides binding site. Notes: The density maps for A2062 and nearby water molecules are contoured at 1.8 in mesh; The E. coli nucleotide numbering is used; the distances between water molecule and nearby atoms are shown in yellow dashed lines. The water molecules are in small red spheres from Mtb structure compared with the corresponding waters in wheat spheres from two structures (PDB codes 6XHX and 6XHY).
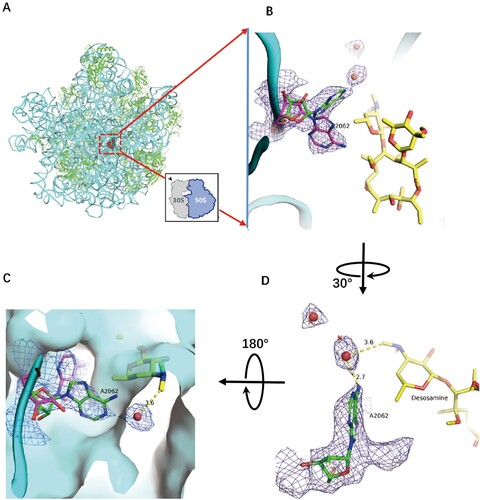
Figure 3. The structure comparison of A2062 in the ribosome complexes with CTY or ERY. (A)(B) The alternative conformations of A2062 with density map contoured at 1.5 from different views (syn-A2062 in Mtb 50S ribosome-CTY complex or CTY-Mtb ribosome complex in green, anti-A2062 in ERY-Eco complex from PDB 4V7U in red; A2062 in ERY-Tth ribosome complex from PDB 6XHX in orange, with 1 Å deviation of base comparing to the anti-A2062 in CTY-Mtb ribosome complex in cyan); (C) The A2062 structure comparison between the non-swayed conformations from different ribosomes (CTY-Dra from PDB 1J5A in pink, ERY-Asu from PDB 6S0X in yellow); (D) The A2062 rotation between two conformations (the rotation about 178°, while the plane angle of two bases is about 66°).
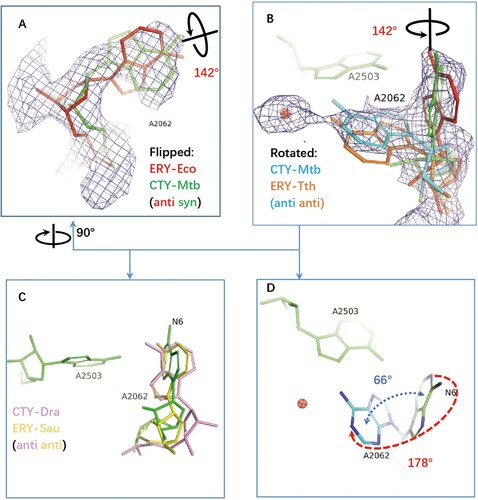
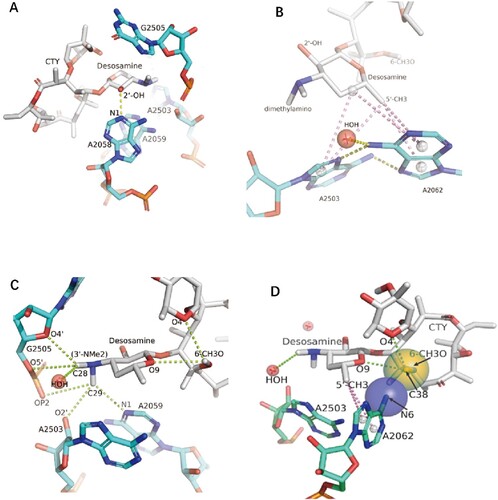
Figure 4. The interactions of CTY with the ribosome. (A)The major nucleotides of the NEPT interacting with CTY from A2058 side view; (B) The π-interactions between desosamine and a Hoogsteen base pair formed by m2A2503 and the swayed A2062; (C) The interactions between Dimethylamino group (3’-NMe2) of desosamine and m2A2503, G2505 and A2059; (D) The interactions of the N6-methoxy Bridge between CTY and the non-swayed A2062 (N6 of A2062 in blue sphere, methylation group of 6-methoxy of CTY in orange sphere); Notes: H-bonds in yellow dotted lines, π-interactions in pink dotted lines, C–H H-bonds in limon dotted lines.