Figures & data
Figure 1. Comparison of the infection features between the Delta and Omicron variants. A. Schematic illustration of the spike mutations of the Omicron variants. Mutations on the Omicron spike protein are indicated by black arrows. SP, signal peptide; NTD, N-terminal domain; RBD, receptor-binding domain; CTD, C-terminal domain; TM, transmembrane domain; CT, cytoplasmic tail. B. Entry efficiency of the SARS-CoV-2 variants in BHK21-hACE2 cells. SARS-CoV-2 pseudoviruses carrying the indicated spike proteins were incubated with the BHK21-hACE2 cells at the indicated TCID50. Luciferase activity was determined at 16 h post-infection (hpi). RLU: Relative Luciferase Unit. Data were presented as Mean ± SD. Statistical significance was determined using unpaired two-tailed t-test, **indicates P < 0.01, ***indicates P < 0.001. C. Entry competition assay of the Delta and Omicron variants in BHK21-hACE2. The indicated Delta and Omicron pseudoviruses carrying either EGFP or Mcherry reporter gene were mixed with the TCID50 = 1:1 and then incubated with the BHK21-hACE2 for co-infection. At 24 hpi, cell nuclei were stained by Hoechst 33342 for 30 mins, and the images were captured by the microscope. Delta and Omicron were indicated as “D” and “O” in the merged channel, respectively. Scale bars, 200 μm. D. Fusion efficiency of different variants’ spikes in Vero-E6 cells. BHK21-hACE2 and Vero-E6 Cells were transfected with plasmid vector or plasmids expressing spike proteins of the WT-D614G, Delta, or Omicron variants. Next, BHK21-hACE2 cells were infected with VSV-ΔG-GFP at 4 h after transfection, Vero-E6 cells were infected 24 h after transfection. At 8 hpi, cells with or without syncytia formations were treated with Hoechst 33342 for nuclei staining (blue). Images of the green channel and the bright field were captured by the microscope. Scale bars, 200 μm.
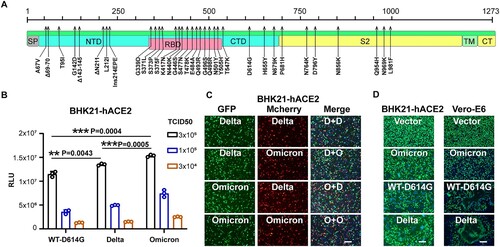
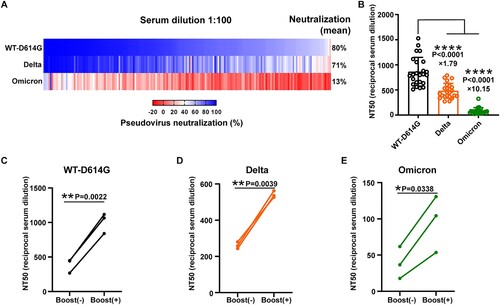
Figure 2. The neutralization efficiency of Wuhan convalescents’ sera against the indicted SARS-CoV-2 variants. A. The heatmap of sera neutralization efficiency against the WT-D614G, Delta, and Omicron variants. Sera from 180 convalescents were 100-fold diluted in the neutralization assay. FBS was used as a negative control. The average sera neutralization efficiency against the WT-D614G, Delta, Omicron were 80%, 71%, 13%, respectively. B. The half neutralization titer of 24 sera against the WT-D614G, Delta, and Omicron variants, respectively. The fold-change of the average NT50 titer was indicated on the data points. Data were presented as Mean ± SD. Statistical significance was determined using a paired two-tailed t-test. ****indicates P < 0.0001. C-E. The effect of vaccination on the sera neutralizing activity against the variants in convalescents. The NT50 titer of the sera collected before or after the vaccine boosting was determined against the WT-D614G(C), the Delta (D), and the Omicron (E).
Data availability
All data are available in the manuscript.
