Figures & data
Figure 1. Transmission chain of a probable pet shop-related outbreak due to SARS-CoV-2 Delta AY.127 variant. Male and female laboratory-confirmed COVID-19 patients linked with this outbreak are represented by blue and red icons, respectively. The patients who had received at least two doses of inactivated or mRNA COVID-19 vaccines are indicated by ticks. The case numbers are represented by the numbers in square brackets above the sex and age of each patient. The transmission chain related to pet shop A involved transmission within family members and dining premises. The transmission chain related to pet shop B was limited. The transmission chain related to pet shop C involved transmission within family members and other residents in the same housing estate A and housing estate B.
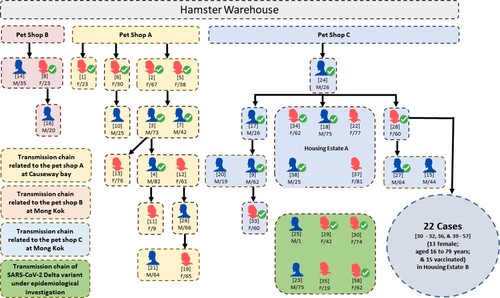
Figure 2. Epidemic curve of a probable pet shop-related outbreak of SARS-CoV-2 Delta AY.127 variant. A total of 58 COVID-19 cases were reported. The timeline represents the date of symptom onset for symptomatic cases or the date of reporting for asymptomatic COVID-19 cases.
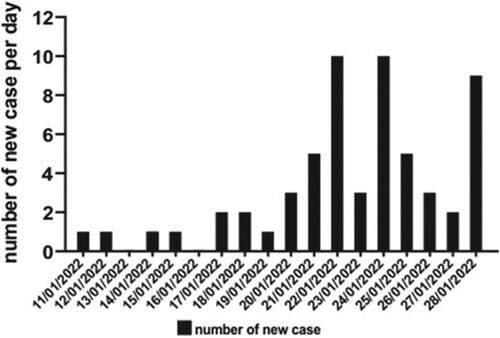
Figure 3. Phylogenetic tree of whole-genome sequences of SARS-CoV-2 found in patients, hamsters or environment of pet shops. Complete viral genome nucleotide sequences were aligned, and the phylogenetic tree was generated by Maximum Likelihood method and generalized time-reversible substitution model GTR + F+I. The number next to the branches indicated the bootstrap values representing the percentage of 1000 replicates. Full-length genomic sequences derived from 15 patient samples and six hamster swab samples, one representative genome sequence of original SARS-CoV-2 (Wuhan_Hu_1), five Variant of Concern strains (Alpha, B.1.1.7; Beta, B.1.351; Gamma, P.1; Delta, B.1.617.2; Omicron, B.1.1.529) and SARS-CoV-2 lineage B.1.258 were included in the analysis.
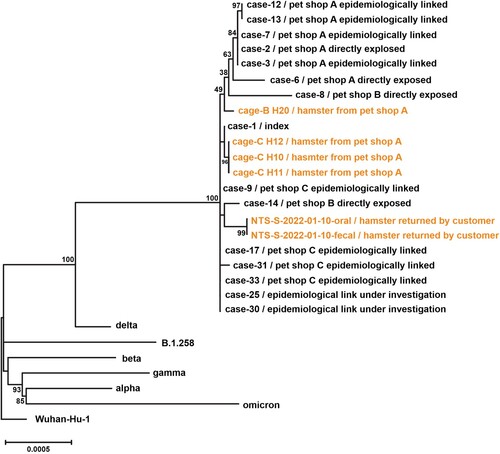
Figure 4. Phylogenetic tree of SARS-CoV-2 spike amino acid sequences from patients, the environment of the pet shop, hamster of a pet shop or wholesale warehouse. Amino acid sequences of spike protein were aligned, and the bootstrap consensus tree was generated by the Neighbor-Joining method. Spike sequences derived from lung tissue of three hamsters (L59, L60 and L64), representative spike amino acid sequence of original SARS-CoV-2 (Wuhan_Hu_1), Variant of Concern strains (Alpha, B.1.1.7; Beta, B.1.351; gamma, P.1; Delta, B.1.617.2; Omicron, B.1.1.529), one closest strain to hamster spike sequences (B.1.258) and spike sequences derived from 12 patient samples and six hamster swab samples were included in the analysis. Detailed information of reference sequences (accession ID, location and virus name) was listed in Supplementary Table 1.
Figure 5. Mutations of pet shop strains and warehouse strains L59, L60 and L64 compared to Wuhan-Hu-1. (A) Sequence alignment of spike protein mutation sites. The corresponding domains of the mutations were indicated. (B) Locations of the RBD mutations in three-dimensional RBD-ACE2 complex structure. ACE2 and RBD were shown in the dark and light grey cartoon, respectively. The mutation sites were highlighted with a red sphere. The predicted side-chain conformations of RBD with mutations were shown in green and cyan stick representation.
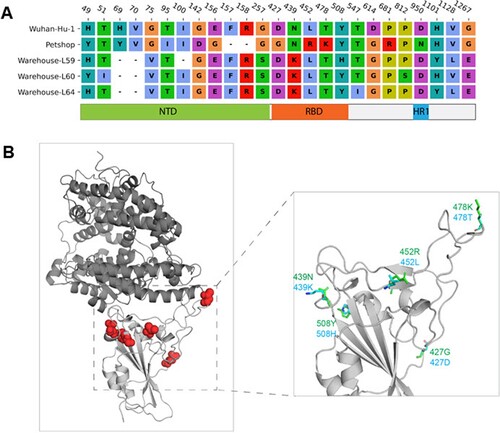
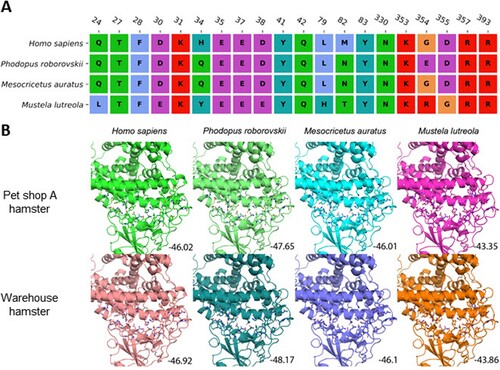
Figure 6. Protein-protein docking between RBD and ACE2. (A) Sequence alignment of ACE2 residues in human (Homo sapiens), dwarf hamster (Phodopus roborovskii), golden Syrian hamster (Mesocricetus auratus) and European mink (Mustela lutreola) interacting with SARS-CoV-2 spike protein RBD. (B) Interactions between ACE2 and SARS-CoV-2 spike RBD from the pet shop and warehouse hamster virus. Interacting amino acids are shown in stick representation. Predicted binding scores are indicated in the bottom corner.