Figures & data
Figure 1. Timeline of the clinical course of the patient and identification of the causative pathogen.
Figure 2. Chest radiographs during the clinical course. Bilateral ground-glass opacity and consolidation were observed on day 9 (Dec 26, Panel A) and worsened on day 11 (Dec 28, Panel B). Bilateral lung involvement was slightly relieved on day 14 (Dec 31, Panel C) after methylprednisolone was administrated on day 11. Bilateral ground-glass opacity and consolidation continued till day 16 (Jan 2, Panel D) and day 18 (Jan 4, Panel E) and relieved slightly on day 20 (Jan 6, Panel F).
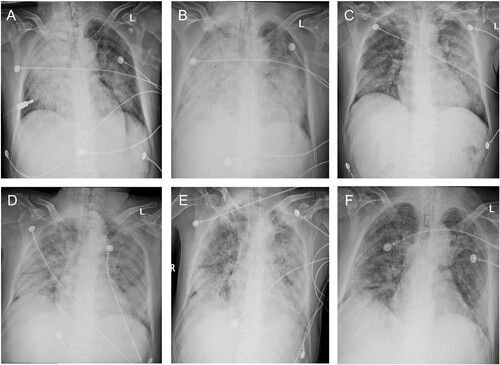
Table 1. Clinical blood biochemistry tests and gas analysis.
Table 2. Complications, treatment, and clinical outcome of the patients.
Figure 3. Metagenomic analysis of the clean reads after host sequences were removed. (A) The complete genome of reference APMV-1 strain pigeon/China/SDLC/2011 was fully covered by 3,289 reads with a mean depth of 31.2×. (B) taxa assignment of the clean reads using MEGAN software. Taxa and the number of their assigned reads were labelled in the polygenetic tree. The pie chart showed Acinetobacter baumannii contributed the most reads, followed by pigeon APMV-1 virus, among the reads of microorganism origin.
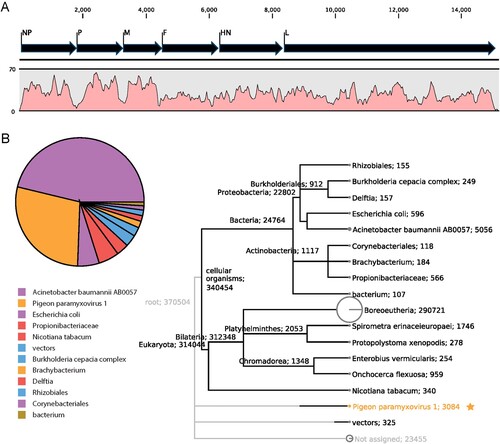
Figure 4. Phylogenetic classification of the APMV-1 virus using the full-length nucleotide sequence of the fusion gene, according to the phylogenetic classification system suggested by Dimitrov et al. [Citation21]. The patient’s APMV-1 virus clustered with pigeon strains isolated in north China and belonged to genotype VI.2.1. Pigeon strains from China were labelled with a green triangle and the patient’s APMV-1 strain was labelled with a red circle. The cleavage sites of the fusion protein were aligned for several strains shown in the bottom-right corner, from which multiple basic amino acids were observed, indicating high virulence of the patient’s strain to poultry. Strain name abbreviations: Human/NY, Avian paramyxovirus 1 from the lethal human case of pneumonia reported by Goebel, et al. [Citation15]; Human/SD, APMV-1 strain from our case occurred in Shangdong province in China; pigeon/1344, pigeon/Qinghai/1344/2017; pigeon/TJ2017, pigeon/China/TJ201; Pigeon/190610, Pigeon/Shandong/190610-2/2019; Pigeon/3P3, Pigeon/Shandong/3P3/2018.
![Figure 4. Phylogenetic classification of the APMV-1 virus using the full-length nucleotide sequence of the fusion gene, according to the phylogenetic classification system suggested by Dimitrov et al. [Citation21]. The patient’s APMV-1 virus clustered with pigeon strains isolated in north China and belonged to genotype VI.2.1. Pigeon strains from China were labelled with a green triangle and the patient’s APMV-1 strain was labelled with a red circle. The cleavage sites of the fusion protein were aligned for several strains shown in the bottom-right corner, from which multiple basic amino acids were observed, indicating high virulence of the patient’s strain to poultry. Strain name abbreviations: Human/NY, Avian paramyxovirus 1 from the lethal human case of pneumonia reported by Goebel, et al. [Citation15]; Human/SD, APMV-1 strain from our case occurred in Shangdong province in China; pigeon/1344, pigeon/Qinghai/1344/2017; pigeon/TJ2017, pigeon/China/TJ201; Pigeon/190610, Pigeon/Shandong/190610-2/2019; Pigeon/3P3, Pigeon/Shandong/3P3/2018.](/cms/asset/7e964661-2095-4b56-90c2-6d03eebd3ec3/temi_a_2054366_f0004_oc.jpg)
Supplemental Material
Download TIFF Image (5.1 MB)Data availability statement
Sequences are available via the National Genomics Data Center (BioProject number PRJCA006351under accession CRA004864).
