Figures & data
Table 1. H5N6 viruses bearing the clade 2.3.4.4b HA gene isolated in this study.
Figure 1. Phylogenetic tree of the HA genes of H5 viruses. The Bayesian time-resolved phylogenetic tree of the clade 2.3.4.4 HA gene of 170 H5 viruses, including the 15 novel H5N6 reassortants in this study and 155 representative viruses reported by others. The phylogenetic tree of the HA gene with more complete information is shown in the Supporting Figure S1 and was rooted to A/goose/Guangdong/1/1996 (H5N1). The viral names of the 59 clade 2.3.4.4b HAs are shown in the small tree, in which the novel H5N6 viruses sequenced in this study are shown in red and marked with red dots. The H5N6 viruses isolated from humans are marked with red solid triangles.
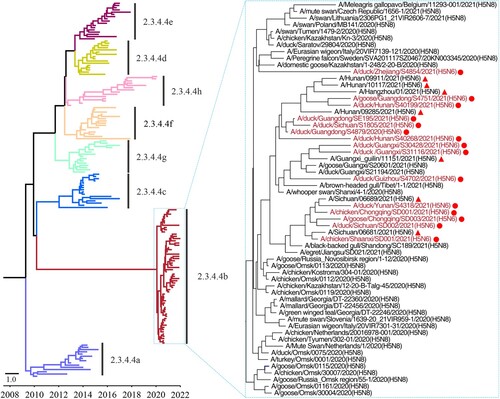
Figure 2. Phylogenetic analyses of the NA gene and the six internal genes of the H5N6 viruses. Phylogenetic analysis was performed by using the MEGA 7.0 software package, implementing the neighbor-joining method. The tree topology was evaluated by 1,000 bootstrap analyses. (a) The phylogenetic tree of the NA genes of 63 H5N6 viruses was rooted to A/Changsha/1/2014 (H5N6). (b-g) The phylogenetic trees of the PB2, PB1, PA, NP, M, and NS genes were rooted to A/goose/Guangdong/1/1996 (H5N1). The novel H5N6 viruses sequenced in this study are shown in red and marked with red dots. The H5N6 viruses isolated from humans are marked with red solid triangles.

Figure 3. Genotypes of the H5N6 viruses and the hosts from which these genotypes were detected. The eight bars represent the eight gene segments (from top to bottom: PB2, PB1, PA, HA, NP, NA, M, and NS), and the colour of the bar indicates the closest donor strain of the gene segment.
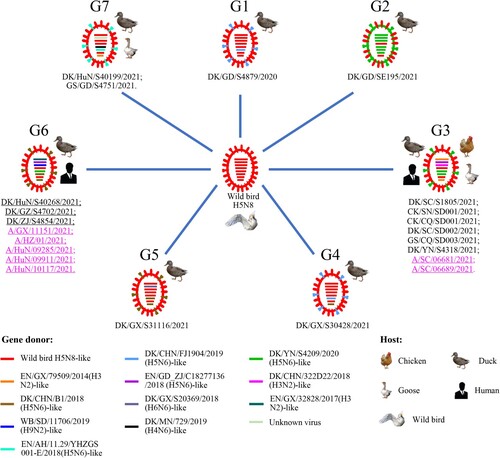
Table 2. Mutations detected in the H5N6 viruses that contribute to the increased binding to human-type receptors and virulence in mammals.
Figure 4. Receptor-binding properties of the H5N6 and H5N8 viruses bearing the clade 2.3.4.4b HA gene. Binding of the indicated viruses to sialylglycopolymers (α-2,3-sialylglycopolymer, blue; α-2,6-sialylglycopolymer, pink). The data shown are the means of three repeats; the error bars indicate standard deviations.
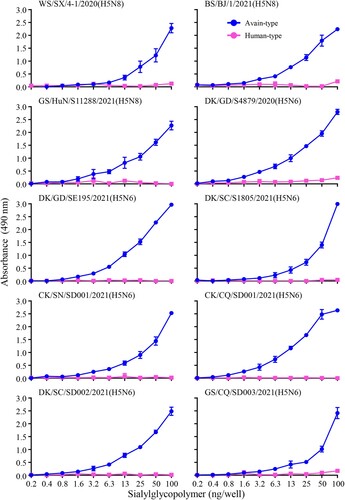
Figure 5. Replication and virulence of H5N6 viruses in mice. (a) Virus titers in organs of mice inoculated intranasally with 106 EID50 of different H5N6 viruses. Three mice from each group were euthanized and their organs were collected on day 3 post-inoculation for virus titration in eggs. Data shown are means ± standard deviations. The dashed lines indicate the lower limit of detection. (b) The death pattern and MLD50 of the indicated viruses.
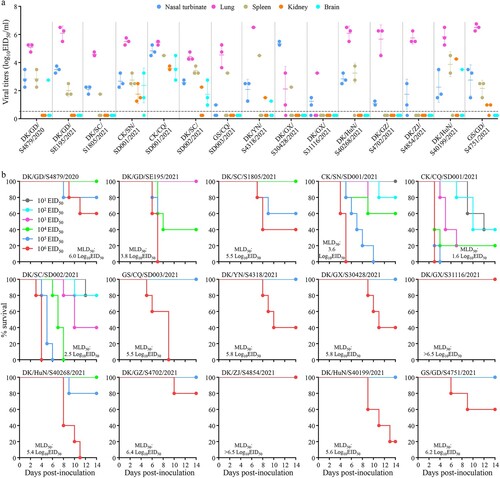
Table 3. Cross-reactive HI antibody titers of the novel H5N6 viruses with antisera induced by different vaccine seed viruses or vaccines.
