Figures & data
Table 1. Characteristics of the patients enrolled in this study.
Figure 2. Mapping of genome assemblies for P. putida, E. coli, and Salmonella generated from ONT MinION reads to the respective reference genomes (P. putida: NC_021505.1, E. coli: NC_000913.3, and Salmonella: NC_003197.2). The mean sequencing coverage for the P. putida, E. coli, and Salmonella genomes assembled in this study was approximately 39.8%, 80.1%, and 57.1%, respectively.
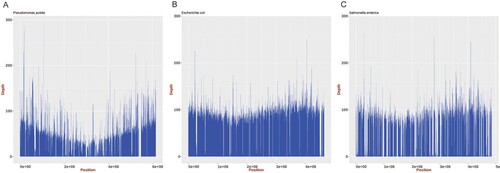
Figure 3. Comparison of the genome sequence of the WGH-01_Typhi isolate obtained in Sierra Leone with other S. Typhi complete genome sequences.
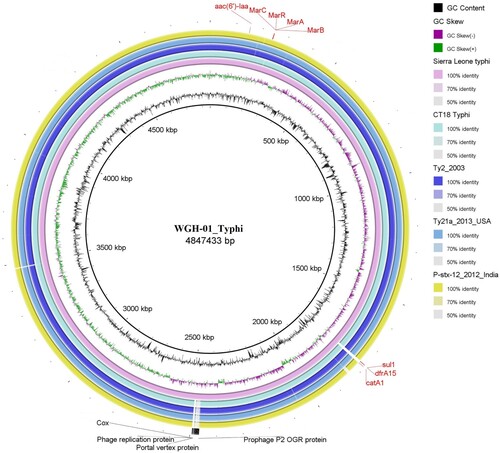
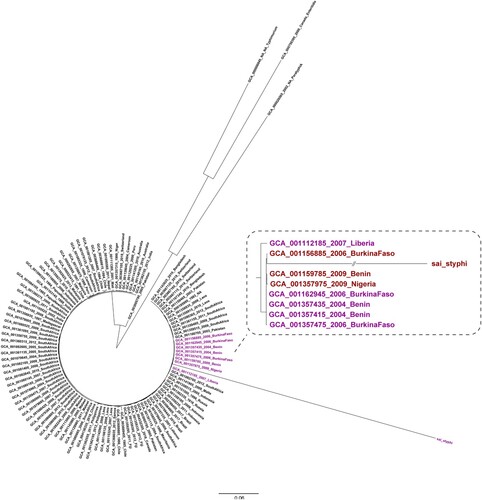
Figure 4. Phylogenetic analysis of the S. Typhi isolate obtained in the present study conducted with sequences from GenBank. The phylogenetic analysis was conducted on the S. Typhi isolate from our study outpatients and on another 105 S. Typhi sequences retrieved from GenBank. The genomes of S. Paratyphi A (GCA000026565), S. Typhimurium (GCA000006945), and S. Enteritidis (GCA000750395) were used as outgroups.