Figures & data
Figure 1. The replicative fitness advantage of Omicron BA.2 over BA.1. (A) Scheme of the in vitro competition model using a mixture of BA.1 and BA.2 variants (final MOI of 0.10 for each variant) in VeroE6-TMPRSS2 and Caco2 cell lines. (B) Scheme of the in vivo competition model using a mixture of both variants with an initial PFU ratio of 1:1 was inoculated in 8–10 weeks old male golden Syrian hamsters (n = 5 per group). At 0 dpi, each hamster was intranasally inoculated with 100 µL of DMEM containing 105 PFU of each variant (n = 5 hamsters per time-point). (C) The BA.2 to BA.1 viral-load ratios in the VeroE6-TMPRSS2 cell lysates were measured by Sanger sequencing at indicated time points. (D) The BA.2 to BA.1 viral-load ratios in the Caco2 cell lysates were measured by Sanger sequencing at indicated time points. (E) The BA.2 to BA.1viral-load ratios of indicated time-point of hamster lung samples were determined by Sanger sequencing. Data in (C–E) are indicated as mean ± SD. P values are calculated as coefficient of each linear regression analysis of indicated time-point ratios versus baseline ratios (1:1). N.S., not significantly different; ***P < 0.001; ****P < 0.0001.
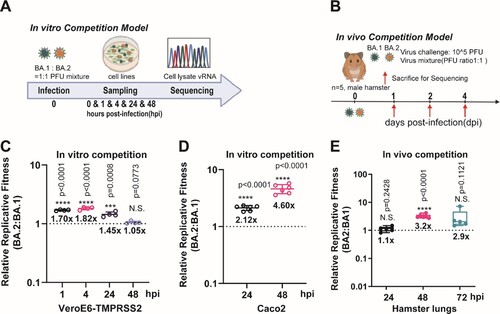
Figure 2. Construction of SARS-CoV-2 mutants and plaque morphologies of different recombinant mutants. (A) Schematic summary of mutations of BA.1 and BA.2 in the spike protein S1 domain. (B) The binding interface between human ACE2 (sand) and SARS-CoV-2 RBD wildtype (G496), S496, G496R498, and S496R498 mutations. Key residues at the binding interface were labelled and shown in stick representation. (C) Construction of revertant SPIKE-G496S (S496), -Q498R (G496R498), and -double mutants (S496R498) SARS-CoV-2 based on the p-BAC backbone of wild-type SARS-CoV-2 (Wuhan/Hu-1/2019). (D) Sanger sequencing of wildtype-backbone (G496), S496, G496R498, and S496R498 mutations. (E) Plaque morphologies of wildtype-backbone (G496), S496, G496R498, and S496R498 mutations. The plaque images were taken on 60hpi and on VeroE6-TMPRSS2 cells.
Figure 3. The spike G496S substitution reduces replicative fitness of Omicron BA.1 against BA.2 (A) In vitro virus competition assay using a mixture of the wildtype (G496) and S496 mutant. The viral-load ratios of G496 to S496 in the cell supernatants were measured by Sanger sequencing. (B) In vivo virus competition assay measuring the wildtype (G496) to S496 viral-load ratios in the nasal turbinate, trachea, and lung of male hamsters (n = 5 hamsters) at 2 dpi by Sanger sequencing. (C) In vitro virus competition assay using a mixture of the G496R498 single mutant and the S496R498 double mutant. The viral-load ratios of G496R498 to the S496R498 in the cell culture supernatants were determined by Sanger sequencing. (D) In vivo virus competition assay measuring the viral-load ratios of G496R498 single mutant to the S496R498 double mutant were performed in male hamsters (n = 5 hamsters) at 2 dpi by Sanger sequencing. All data are indicated as mean ± SD. P values are calculated as the coefficient of each linear regression analysis of indicated group ratios versus baseline ratios (1:1). **P < 0.01; ***P < 0.001; ****P < 0.0001.
Figure 4. mAb neutralizing activity against S496 and G496 variants. Pairwise neutralizing antibody titres (half-maximum inhibitory dose; ID50) against S496 single mutant and G496 wildtype. The mAbs are divided into two classes according to their opposite phenotypes of antibody evasion. The result is shown as mean ± SD of triplicate and repeated twice for confirmation. Data is analysed by unpaired t test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.

