Figures & data
Table 1. Average results in accuracy terms by repeating 20 times a 10-fold cross validation analysis 20 times for all ML supervised algorithms for each method and step.
Figure 1. Distance plot of RF and PLS-DA for discrimination between ESRI developers susceptible to P/T and their isogenic isolates resistant to P/T. A. C, Full Spectrum method. B. D, Threshold method. ESRI: Extended spectrum resistance to BL/BLI susceptible to P/T; Pressed: ESRI isogenic isolates resistant to P/T after pressure with P/T; PLS-DA: partial least squares discriminant analysis; RF: random forest. t0 and t1 are two components of an algorithm called MDS that help to represent data from a distance matrix.
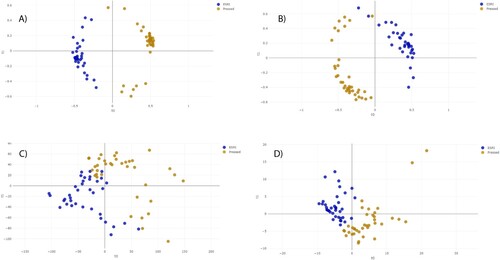
Figure 2. Distance plot of RF and PLS-DA for discrimination between P/T-susceptible and P/T-resistant E. coli (first step analysis). A. C, Full Spectrum method. B. D, Threshold method. PLS-DA: Partial Least Square Discriminant Analysis; RF: Random Forest. t0 and t1 are two components of an algorithm called MDS that help to represent data from a distance matrix.
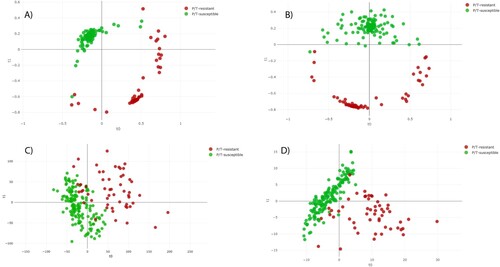
Figure 3. Distance plot of RF and PLS-DA for discrimination between P/T-susceptible and ESRI developer Escherichia coli (second step analysis). A. C, Full Spectrum method. B. D, Threshold method. PLS-DA: Partial Least Square Discriminant Analysis; RF: Random Forest. t0 and t1 are two components of an algorithm called MDS that help to represent data from a distance matrix.
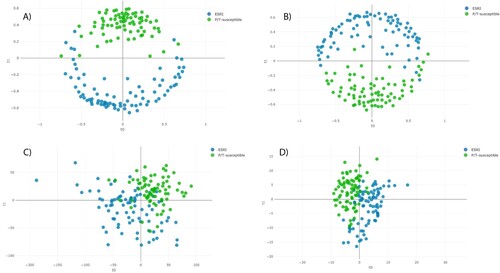
Figure 4. Feature importance from RF analysis from each assay. A, ESRI developers susceptible to P/T and their isogenic isolates resistant to P/T. B, P/T-susceptible and P/T-resistant E. coli (Step 1). C, P/T-susceptible and ESRI developers susceptible to P/T (Step 2). RF: Random Forest; ESRI: Extended Spectrum Resistance to BL/BLI susceptible to P/T.
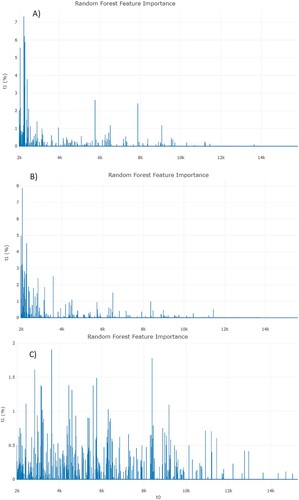
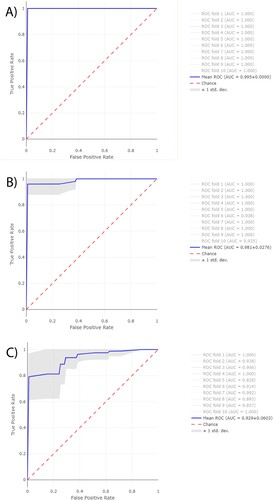
Figure 5. Average ROC curves from first 10-folds and their AUC values obtained from RF using Full Spectrum method to discriminate between A, ESRI developers susceptible to P/T and their isogenic isolates resistant to P/T B, P/T-susceptible and P/T-resistant E. coli and C, P/T-susceptible and ESRI developer Escherichia coli. Isogenic isolates resistant to P/T, resistant isolates to P/T and ESRI developer isolates susceptible to P/T were labelled as positive categories respectively.
Supplemental Material
Download MS Word (30.4 KB)Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.