Figures & data
Figure 1. Isolation of monoclonal antibodies from single B cells of convalescent COVID-19 patients. (A) RBD-specific binding activities of sera derived from 3 (P1–P3) convalescent and 1 (P4) acute COVID-19 patients as measured by ELISA; (B) Spike-specific binding activities of sera derived from four COVID-19 patients as measured by ELISA; (C) Neutralization activities of sera derived from four COVID-19 patients as measured by pseudotyped SARS-CoV-2 inhibition in 293T-ACE2 cells; (D) Antibody gene repertoire analysis of reactive B cells derived from each patient. The number of cloned antibody genes from each patient is shown in the center of each pie chart for both the heavy (H) and light (L) chains. The colors represent specific variable gene family. Each fragment of the same color stands for one specific sub-family; (E). The percentage of somatic hypermutation (SHM) compared to germline sequences and the CDR3 amino acid lengths of cloned antibody H and L gene sequences were analyzed for each subject; (F) RBD (left) and spike (right) specific binding activities of five HuNAbs, including A6, B4, B7, B8 and C5, were measured by ELISA; (G) Neutralization activities of 5 HuNAbs against pseudotyped (left) and authentic (right) SARS-CoV-2 were determined in HEK 293T-ACE2 and Vero-E6 cells, respectively. HIV-1 specific HuNAb VRC01 served as a negative control. Each assay was performed in duplicates and the mean of replicates is shown with the standard error of mean (SEM); (H) The competition of four HuNAbs, including B4, B7, B8 and C5, with human soluble ACE2 for binding to SARS-CoV-2 RBD was measured by SPR. The curves show binding of ACE2 to SARS-CoV-2 RBD with (red) or without (black) pre-incubation with each HuNAb.
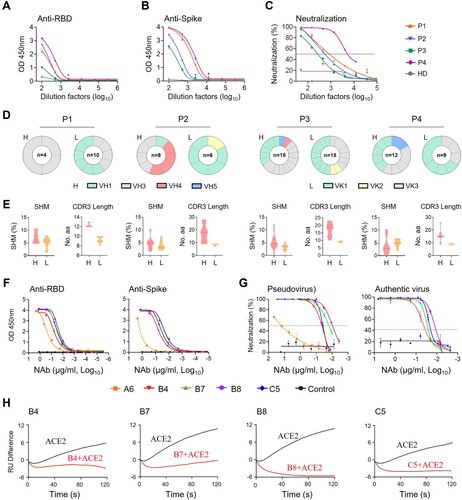
Figure 2. Pre- and post-exposure treatment of B8-IgG1 against SARS-CoV-2 in Syrian Hamster. (A) Experimental schedule. Four groups of hamsters (G1-G4) received intraperitoneally a single dose of 1.5 mg/kg of B8-IgG1 at one day before infection (−1 dpi) for pre-exposure prophylaxis, and at day one (1 dpi), two (2 dpi) and three (3 dpi) post-infection for early treatment, respectively. Control hamsters (G0, n = 4) received an isotypic control antibody at the same dose. On day 0, each hamster was intranasally challenged with a dose of 105 PFU of SARS-CoV-2 (HKU-001a strain). All hamsters were sacrificed on 4 dpi for analysis; (B) Infectious virus (PFU) was measured in animal lungs by the viral plaque assay in Vero-E6 cells. The PFU/ml concentration is shown in log-transformed units; (C) The relative viral RdRp RNA copies (normalized to β-actin) were determined by RT-PCR in animal lungs; (D) Representative 100× images of infected lungs from each group, as determined by anti-NP immunofluorescence (IF) staining. The cell nuclei were counterstained with DAPI (blue); (E) Infectious virus (PFU) were measured in NT homogenates by the viral plaque assay as mentioned above; (F) Viral loads in NT homogenates of each group were determined by RT-PCR assay. The viral load data is shown in log-transformed units; (G) Representative 100× images of infected NT from each group as determined by anti-NP IF staining as mentioned above. Statistics were generated using one-way ANOVA tests. *p < 0.05; **p < 0.01; ***p < 0.001.
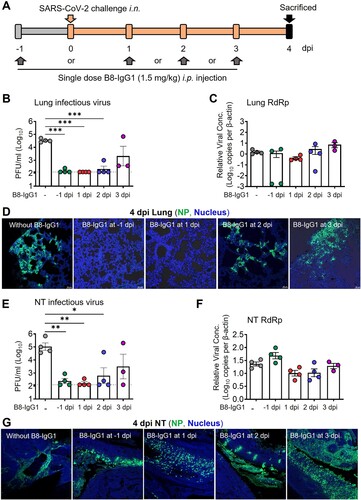
Figure 3. Pre-exposure treatment with monomeric B8-mIgA1 or B8-mIgA2 against SARS-CoV-2 infection in Syrian hamsters. (A) Experimental schedule. Five groups of hamsters (n = 4 per group) received a single dose of 4.5 mg/kg of B8-IgG1, B8-mIgA1 or B8-mIgA2 one day before viral challenge for pre-exposure prophylaxis by the intranasal route (circles) or the intraperitoneal route (triangles), respectively. Control hamsters (n = 4) received PBS. On day 0, each hamster was intranasally challenged with a dose of 105 PFU of SARS-CoV-2, as mentioned in A. All hamsters were sacrificed on 4 dpi for analysis; (B) The viral RNA load, measured by relative RdRp RNA copy numbers (normalized to β-actin) was determined by RT-PCR in animal lung homogenates; (C) The relative sub-genomic nucleocapsid (sgNP) RNA copy numbers (normalized to β-actin) were determined by RT-PCR in animal lung homogenates; (D) Infectious virus (PFU) was measured in animal lung homogenates by the viral plaque assay in Vero-E6 cells; (E) Representative lung images of infected animals by scanning whole tissue section. The signal of SARS-CoV-2 NP was shown in bright spots; (F) The relative viral RdRp RNA copy numbers (normalized to β-actin) were determined by RT-PCR in NT homogenates; (G) The relative sgNP RNA copy numbers (normalized to β-actin) were determined by RT-PCR in NT homogenates; (H) Infectious virus (PFU) was measured in animal NT homogenates by the viral plaque assay in Vero-E6 cells; (I) Representative NT images of infected animals by scanning whole tissue section. The signal of SARS-CoV-2 NP was shown in bright spots. Log-transformed units are shown in (B) to (H) except in (E). Statistics were generated using one-way ANOVA tests. *p < 0.05; **p < 0.01.
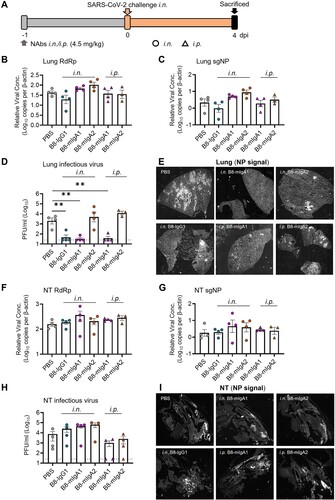
Figure 4. Pre-exposure treatment with dimeric B8-dIgA enhances SARS-CoV-2 infection in Syrian hamsters. (A) Experimental schedule. Four groups of hamsters (n = 4 per group) were inoculated intranasally with B8-dIgA1 or B8-dIgA2, either at a low dose of 4.5 mg/kg or at a high dose of 13.5 mg/kg, respectively, 12 hours before intranasal viral challenge. Another group of hamsters (n = 4) received PBS as control. On day 0, each hamster was intranasally challenged with a dose of 105 PFU of SARS-CoV-2 as described in A. All hamsters were sacrificed on 4 dpi for analysis. Data represent a presentative experiment from three independent experiments; (B) The relative viral RdRp RNA copy numbers (normalized to β-actin) were determined by RT-PCR in animal lung homogenates; (C) The relative viral sgNP RNA copy numbers (normalized to β-actin) were determined by RT-PCR in animal lung homogenates; (D) Infectious virus (PFU) was measured in animal lung homogenates by the viral plaque assay in Vero-E6 cells; (E) The relative viral RdRp RNA copy numbers (normalized to β-actin) were determined by RT-PCR in NT homogenates; (F) The relative viral sgNP RNA copies (normalized to β-actin) were determined by RT-PCR in NT homogenates; (G) Infectious virus (PFU) was measured in NT homogenates by the viral plaque assay in Vero-E6 cells; (H) SARS-CoV-2 infection results in more extensive and severe damage of the NT epithelium in B8-dIgA-administrated animals (10× HE images). Injury of the NT epithelium is extensive with partial (black asterisk) or complete (blue asterisk) desquamation in B8-dIgA1- and B8-dIgA2-pretreated Syrian hamsters; (I) Representative lung images of infected animals by scanning whole tissue section. The signal of SARS-CoV-2 NP was shown in bright spots; (J) Representative NT images of infected animals by scanning whole tissue section. The signal of SARS-CoV-2 NP was shown in bright spots. Log-transformed units are shown in (B) to (H) except in (E). Statistics were generated using one-way ANOVA tests. *p < 0.05; **p < 0.01; (K) Experimental schedule. Two groups of hamsters (n = 4 per group) were inoculated intranasally with 13.5 mg/kg of ctrl dIgA1 or TH335 dIgA1 at 12 hours before intranasal viral challenge. On day 0, each hamster was intranasally challenged with a dose of 105 PFU of SARS-CoV-2 as described in A. All hamsters were sacrificed on 4 dpi for analysis. Data represent a presentative experiment from three independent experiments; (L-N) The viral RNA and infectious virus titer were determined as above in animal lung homogenates; (P-R) The viral RNA and infectious virus titer were determined as above in animal NT homogenates; (O) Representative lung images of infected animals by scanning whole tissue section. The signal of SARS-CoV-2 NP was shown in bright spots; (S) Representative NT images of infected animals by scanning whole tissue section. The signal of SARS-CoV-2 NP was shown in bright spots.
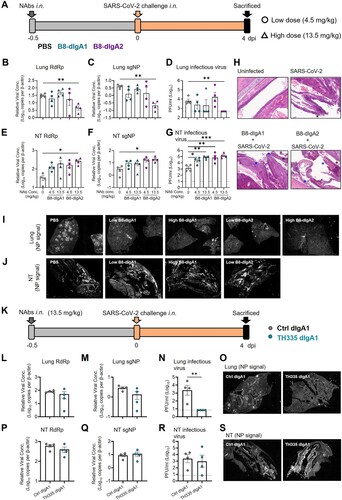
Figure 5. B8-dIgA1 and B8-dIgA2 enhance SARS-CoV-2 infection via CD209. (A) The effects of B8-dIgA2 on SARS-CoV-2 infection in the MucilAirTM model, consisting of primary human nasal epithelial cells but no DCs. B8-mIgA2 or B8-dIgA2 were pre-incubated at doses of 10, 100, and 1000 ng/ml, respectively, in the apical compartment with or without mucus for 1 hour, before adding 104 PFU of SARS-CoV-2 (BetaCoV/France/IDF00372/2020) for 4 hours. The viral RNA loads were measured by RT-PCR in both the apical and basal compartments and are shown in log-transformed units; (B) Representative confocal images (400×) of olfactory epithelium in NT showed the expression of CD209 (DC-SIGN) in green and ACE2 in magenta by immunohistochemical staining of experimental hamsters treated with B8-dIgA2 without (left) or with (middle and right) SARS-CoV-2 infection. Color-coding indicates specific antibodies used for double staining. Infected CD209+ cells are visualized in yellow as indicated by arrows (right); (C) The CD209 or CD299 overexpressed-HEK 293 T cells were pre-treated for 6 hours with 10 ng/ml of B8-dIgA1 or B8-dIgA2 or control dIgA1 or control dIgA2 or PBS, respectively, prior to SARS-CoV-2 infection (MOI: 0.05). Each treatment was performed in triplicate. Two days after infection, SARS-CoV-2 NP expression (green) was quantified by the mean fluorescence intensity (MFI) after anti-NP IF staining. Statistics were generated using student-t tests. *p < 0.05; **p < 0.01; ***p < 0.001L; (D) The effects of B8 antibodies on cell-cell fusion. 293 T cells co-transfected with SARS-CoV-2 spike and GFP were pre-treated with 100× the IC90 dose of B8-IgG1, B8-mIgA1, B8-mIgA2, B8-dIgA1, B8-dIgA2 or and IgG isotypic control for 1 hour, respectively. Vero-E6 cells transfected with TMPRSS2 were then added to the treated 293T-spike-GFP cells and co-cultured for 48 hours. Cell-cell fusion was imaged under a fluorescence confocal microscope at the 50× magnification.
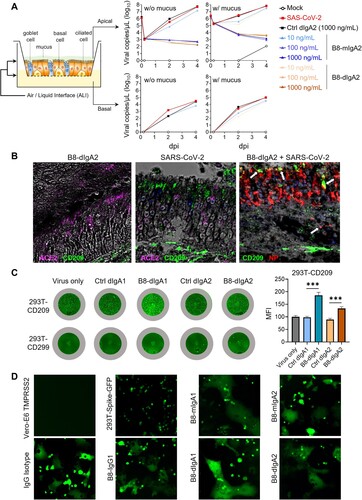
Figure 6. B8-dIgA1 promotes MoDC-mediated transmission of SARS-CoV-2 pseudovirus via CD209. Human MoDCs were treated with 25 ng/ml of different hIgAs for 1 hour. (A) MoDCs were then subjected to flow cytometry analysis. Representative plots showing hIgA-expressing live MoDCs (left) and the expression of CD209 on hIgA positive cells (right). (B) Quantified results depict the frequencies of hIgA positive MoDCs. (C) hIgA-treated MoDCs were subjected to confocal imaging with 20× objective. CD209: green, human IgAs (red) and nucleus (blue). Scale bar: 10 µm. (D) Schedule of cell-to-cell transmission assay. MoDCs were pre-treated with different hIgAs (1) prior to SARS-CoV-2 pseudovirus infection (2). Infected MoDCs were co-cultured with Vero-E6-TMPRSS2 cells (3). (E) Luciferase activity of infected MoDCs with without hIgAs treatments. Luciferase activity of co-culture of MoDCs and Vero-E6 TMPRSS2 with hIgA1 (F) and hIgA2 (G) treatments. Each symbol represents an individual donor with a line indicating the change after different hIgAs treatments.
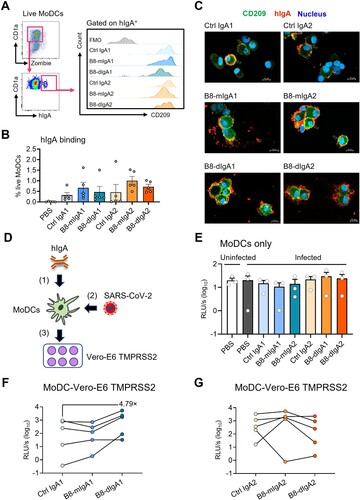
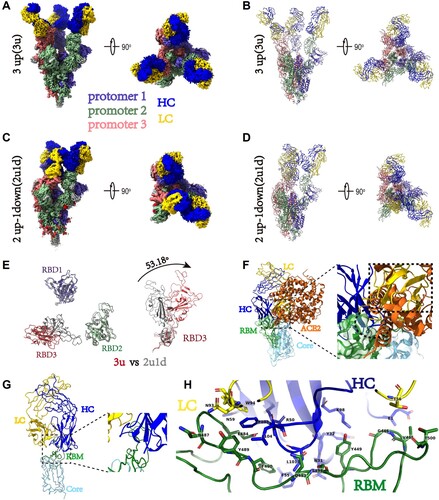
Figure 7. Cryo-EM structures of the SARS-CoV-2 S trimer in complex with B8-Fabs. (A) Side and top views of the Spike-B8 3u cryo-EM map showing 3 up RBDs each bound with a B8 Fab. Protomer 1, 2, and 3 are shown in slate blue, dark sea green, and India red, respectively. Heavy chain and light chain of the B8-Fab are in blue and gold, respectively. This color scheme was used throughout panels (A)-(E); (B) Side and top views of the Spike-B8 3u cryo atomic model; (C) Side and top views of the Spike-B8 2u1d cryo-EM map showing two up RBDs up (RBD-1 and RBD-2) and one RBD down (RBD-3), each bound to a B8-Fab; (D) Side and top views of the Spike-B8 2u1d cryo atomic model. (E) Structural comparation of RBDs between Spike-B8 3u (different colors) and Spike-B8 2u1d (gray); (F) ACE2 (chocolate color, PDB: 6M0J) may clash with the heavy chain (blue) and light chain (gold) of the B8-Fab. ACE2 and the Fab share overlapping epitopes on the RBM (dotted black circle), and the framework of the B8-VL appears to clash with ACE2 (dotted black frame). The RBD core and RBM are shown in light sky blue and green, respectively; (G) Atomic model of an RBD-B8 complex portion in cartoon mode, shown with the same color scheme as in (F); (H) The residues involved in interactions between B8 and the RBM. The heavy and light chain of the B8-Fab are in blue and gold, respectively. The RBM is shown in green.
Supplemental Material
Download MS Word (49.6 KB)Supplemental Material
Download JPEG Image (693.7 KB)Supplemental Material
Download JPEG Image (668.5 KB)Supplemental Material
Download JPEG Image (533.8 KB)Supplemental Material
Download JPEG Image (809.2 KB)Supplemental Material
Download JPEG Image (1.3 MB)Supplemental Material
Download JPEG Image (2 MB)Supplemental Material
Download JPEG Image (1.2 MB)Supplemental Material
Download JPEG Image (1.1 MB)Supplemental Material
Download JPEG Image (987 KB)Data availability statement
The data of this studies are available upon reasonable request and no custom computer code or algorithm used to generate results that are reported in the paper and central to its main claims.
