Figures & data
Figure 1. The workflow of the study. It contains three sections including Data collection, Data analysis and Database construction. Please see the maintext for the details about these sections.
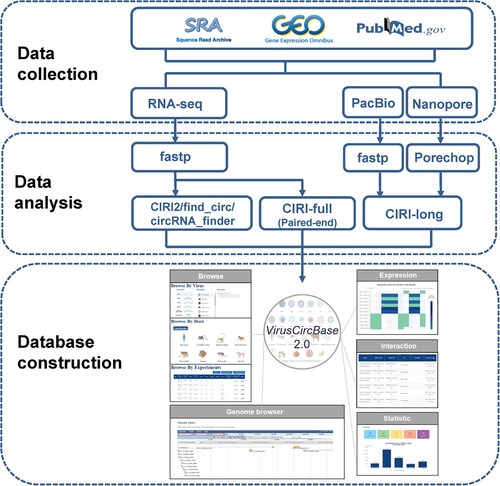
Table 1. Comparison between VirusCircBase 1.0 and VirusCircBase 2.0.
Figure 2. The number of viral circRNAs detected in each virus in the VirusCircBase 2.0. The bar above the x-axis represents the number of circRNAs identified for each virus, with the red portion indicating the newly added viral circRNAs of VirusCircBase 2.0. The first bar below the x-axis represents the Baltimore group to which the virus belongs, while the second bar represents the host kingdom of the virus. For clarity, the abbreviation of virus names was used. Supplementary Table S2 displays the full names of these viruses. ssRNA(+), positive-sense single-stranded RNA virus; dsDNA, double-stranded DNA virus; ssRNA(-), negative-sense single-stranded RNA virus; dsDNA-RT, double-stranded DNA reverse transcribing virus; ssDNA, single-stranded DNA virus; dsRNA, double-stranded RNA virus.
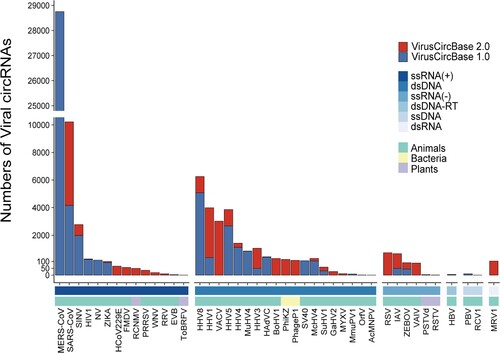
Figure 3. The expression of viral circRNA across different tissues or cells in different hosts. The x-axis represents the abbreviations of different viral species, and the bar above represents the Baltimore classification of the virus. The y-axis represents the host tissues or cells, with the first bar on the left represents the host kingdom and the second bar represents the host species. Each coloured block represents the median expression of viral circRNA in that tissue, with blue and red colours representing low and high expression, respectively, according to the figure legend.
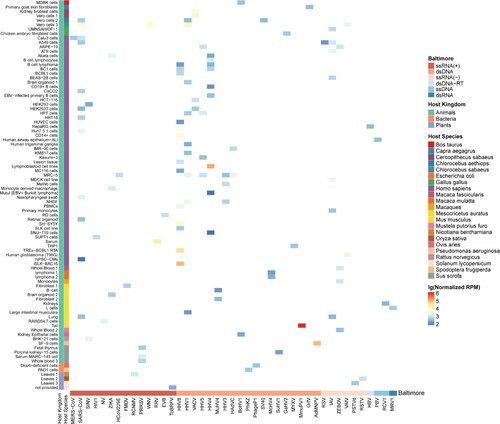
Figure 4. Analysis of the mechanism of viral circRNA formation. (A) Composition of different types of back-splicing signals of viral circRNAs. (B) Composition of different alternative splicing types of viral circRNAs. A5SS, Alternative 5’ splice site; ES, Exon skipping; A3SS, Alternative 3’ splice site; IR, Retained intron. (C) The distribution of the number of isoforms in virus circRNAs.
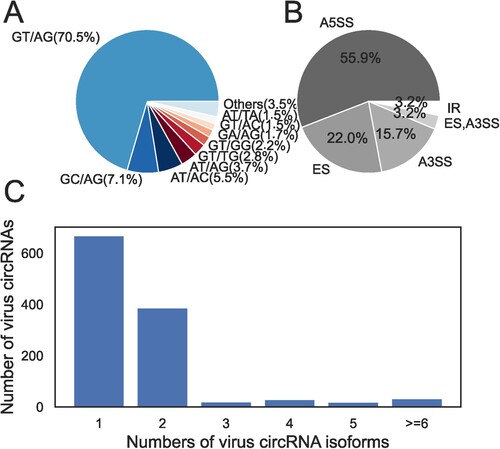
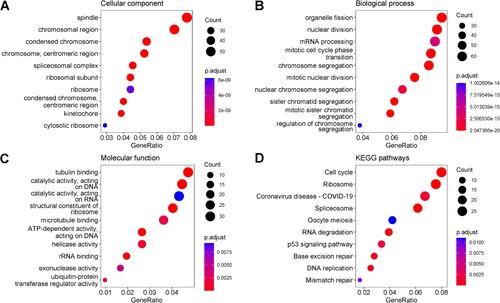
Figure 5. The top 10 enriched GO terms in the domain of Cellular Component (A), Biological Process (B), Molecular Function (C) and the top 10 enriched KEGG pathways (D) for the human genes which had moderate negative correlations with virus circRNA production. The size of dots represented the number of genes in each domain, and the colour indicated the significance of enrichment (adjusted p-values less than 0.05).
Supplemental Material
Download MS Word (904.5 KB)Supplemental Material
Download MS Excel (12.4 KB)Supplemental Material
Download MS Excel (62 KB)Data availability statement
All data used in the study are available in Supplementary materials and in VirusCircBase 2.0 which is accessible at http://computationalbiology.cn/VirusCircBase2/#/home.
