Figures & data
Figure 1. The percentage and frequency of SNVs in brain and placental samples from fetuses infected with the wild Asian-lineage Zika virus strain PRVABC59. (A) The mean percentage (from two technical replicates) of Zika genomic SNVs in the virus stock, fetal brain, and placental samples was calculated from the total number of sequenced virus genome nucleotides. (B) The mean frequency (from two technical replicates) and subtype of Zika SNVs in virus stock, fetal brain, and placental samples. Solid lines represent mean values. Raw data are shown in Table S3-A.
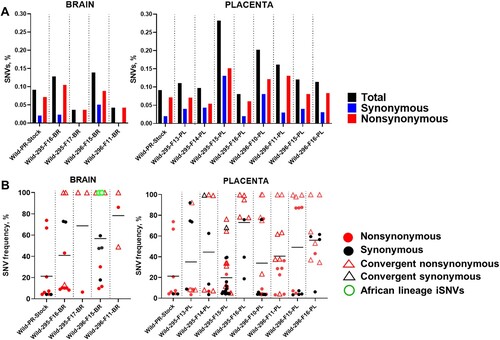
Figure 2. Zika virus iSNVs in the fetal brain and placenta (the wild Asian-lineage Zika virus strain PRVABC59). Patterns of Zika virus iSNVs in brain and placental tissue samples from individual fetuses. C: Zika virus genomic region encoding capsid protein; prM: precursor membrane protein; E: envelope protein; NS1: nonstructural protein 1; NS2A: nonstructural protein 2A; NS2B: nonstructural protein 2B; NS3: nonstructural protein 3; NS4A: nonstructural protein 4A; NS4B: nonstructural protein 4B; NS5: nonstructural protein 5. UTR: untranslated region. Raw data are shown in Table S3-A.
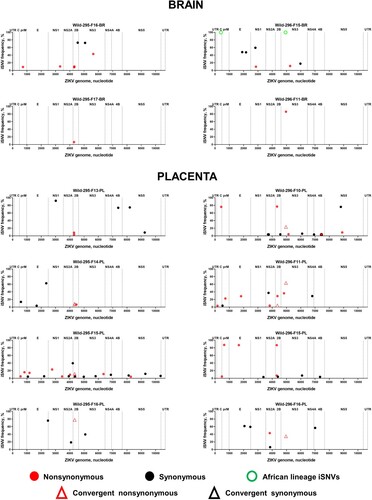
Figure 3. Generic African-lineage Zika virus mutations. Amino acid motif sequences representing the generic African lineage mutations identified in the fetal brain infected with the Asian Zika virus. X-axis – amino acid position in the Zika virus polyprotein.
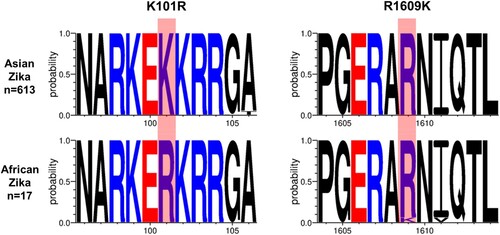
Figure 4. In vitro infection kinetics of WT and AFM Zika virus strains. All cell lines were inoculated with an MOI 0.01. (A) The 24-well plates with cell monolayers were fixed at 72 h after virus inoculation and stained with Zika virus-specific antibodies, and infected cells were counted in the whole 5 wells with a bright-field microscope. Dots represent the number of cells in the individual well. Columns and whisks represent the mean ± SD. (B) Cell culture supernatants were collected at 72 h after virus inoculation, and Zika RNA loads were measured using RT-qPCR. (C) In vitro infection kinetics of WT and AFM Zika virus strains in porcine Dulac cells at 37°C and 39°C. The dotted horizontal line represents the limit of detection. WT: The wild-type Asian Zika virus variant synthetically rescued with Infectious Subgenomic Amplicons. AFM: The fetal brain-specific Zika virus variant synthetically rescued with Infectious Subgenomic Amplicons. *: P < 0.05; **: P < 0.01.
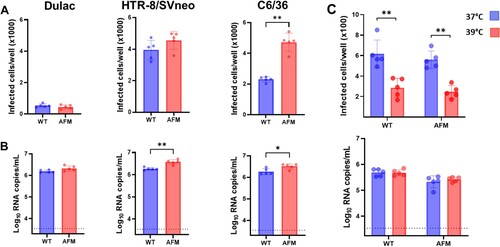
Figure 5. Zika virus loads in the placenta, amniotic fluid, and fetal lymph nodes. Dotted lines represent the limit of detection. Solid lines represent mean values. Solid green and red circles indicate samples from fetuses directly injected with Zika virus. WT: The wild-type Asian Zika virus variant synthetically rescued with Infectious Subgenomic Amplicons. AFM: The fetal brain-specific Zika virus variant synthetically rescued with Infectious Subgenomic Amplicons.
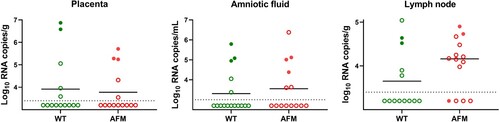
Figure 6. Cytokine levels in the fetal blood plasma. IFN-α, IL-6, IL-8, IL-12, IL-13, and IL-17A in blood plasma were detected in the Bioplex cytokine assay. Solid lines represent means. The dotted line represents the limit of quantification. Solid green and red circles indicate samples from fetuses directly injected with Zika virus. WT: The wild-type Asian Zika virus variant synthetically rescued with Infectious Subgenomic Amplicons. AFM: The fetal brain-specific Zika virus variant synthetically rescued with Infectious Subgenomic Amplicons. *: p < 0.05; **: p < 0.01; ***: p < 0.001; **** p < 0.0001. Control blood plasma samples were from the fetuses of two healthy pigs sampled at 78 days of pregnancy (28 days after in utero inoculation with virus-free media) in our previous study [Citation18].
![Figure 6. Cytokine levels in the fetal blood plasma. IFN-α, IL-6, IL-8, IL-12, IL-13, and IL-17A in blood plasma were detected in the Bioplex cytokine assay. Solid lines represent means. The dotted line represents the limit of quantification. Solid green and red circles indicate samples from fetuses directly injected with Zika virus. WT: The wild-type Asian Zika virus variant synthetically rescued with Infectious Subgenomic Amplicons. AFM: The fetal brain-specific Zika virus variant synthetically rescued with Infectious Subgenomic Amplicons. *: p < 0.05; **: p < 0.01; ***: p < 0.001; **** p < 0.0001. Control blood plasma samples were from the fetuses of two healthy pigs sampled at 78 days of pregnancy (28 days after in utero inoculation with virus-free media) in our previous study [Citation18].](/cms/asset/7a32f74c-0279-4bed-9ad3-77e6ac772978/temi_a_2263592_f0006_oc.jpg)
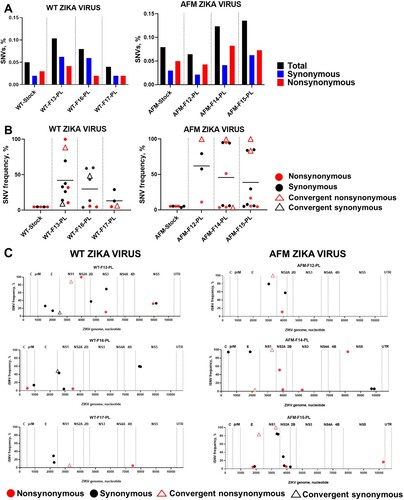
Figure 7. Zika virus SNVs and iSNVs in placental samples from fetuses infected with the wild-type Asian (WT) Zika variant or fetal brain-specific variant with generic African-lineage mutations (AFM). (A) The mean percentage (from two technical replicates) of Zika genomic SNVs in the virus stock and placental samples was calculated from the total number of sequenced virus genome nucleotides. (B) The mean frequency (from two technical replicates) and subtype of Zika SNVs in virus stock and placenta. Solid lines represent mean values. Raw data are shown in Table S3-B, C. (C) Zika virus iSNVs in the fetal placenta. C: Zika virus genomic region encoding capsid protein; prM: precursor membrane protein; E: envelope protein; NS1: nonstructural protein 1; NS2A: nonstructural protein 2A; NS2B: nonstructural protein 2B; NS3: nonstructural protein 3; NS4A: nonstructural protein 4A; NS4B: nonstructural protein 4B; NS5: nonstructural protein 5. UTR: untranslated region. Raw data are shown in Table S3-B, C.
Supplementary_Materials_and_Methods
Download MS Word (103.5 KB)File_S1_Zika_virus_strains
Download MS Word (40.8 KB)File_S2_ISA_DNA_fragments
Download MS Word (25.4 KB)Table_S1_WT_and_AFM_infected_samples_and_NGS_overview
Download MS Excel (19.4 KB)Table_S2_Zika_virus_PrimalSeq_primers
Download MS Excel (14.1 KB)Table_S3_SNVs_and_iSNVs
Download MS Excel (59.2 KB)Table_S4_Primers_for_ISA
Download MS Excel (11 KB)Table_S5_Comparative_in_utero_and_field_Zika_virus_evolution
Download MS Excel (35.6 KB)Data availability
An accession number for raw NGS data is PRJNA837638 in NCBI BioProject.
