Figures & data
Figure 1. Time-calibrated phylogeny of a subset of the global dengue 1 virus genomes alongside the Cameroon 2017–2018 outbreak strains (in red text). Coloured circles indicate geographic origins. Dates of the most recent common ancestor (TMRCA) for major clades are provided in the table inset. Posterior probabilities are shown at major nodes. Taxon labels include GenBank accession number, country, and year of isolation for reference sequences, while new sequences are labelled with strain names. nCAC: new Central African clade; oCAC: old Central African clade.
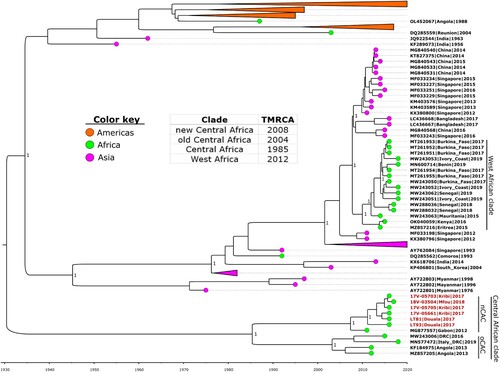
Table 1. Demographics and clinical background of the positive samples.
Table 2. Amino acid substitutions in ORFs of nCAC.
Figure 2. Molecular phylogenetic analysis of a subset of global DENV-1 partial E sequences alongside the Cameroon 2017–2018 outbreak strains (in red text). Tree model inference and phylogeny were simultaneously conducted in IQ-TREE v1.6.1, executing 1000 bootstrap replicates. The geographical origins of the major African clusters are colour coded and indicated in the inset map. Critical nodes are labelled with bootstrap values. The tree was visualized in FigTree v1.4.4.
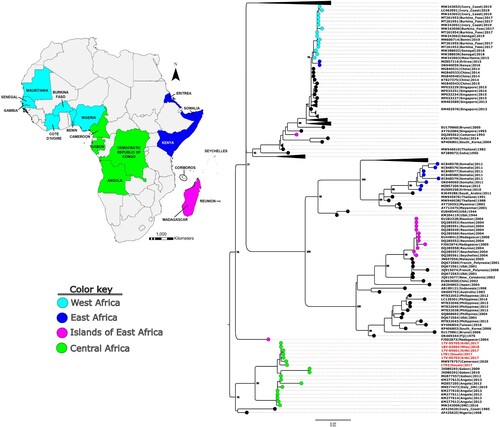
Figure 3. Dengvaxia® and TetraVax-DV-TV003 vaccine strain divergence analysis. The analysis was restricted to the immuno-protective pre-membrane (prM) and the envelope (E) protein region of CYD1. Numerals indicate amino acid positions. Only amino acid positions with disagreements are shown. Single-point disagreements are highlighted. CYD1: Dengvaxia®; TV003: TetraVax-DV-TV003; nCAC: new Central African clade; oCAC: old Central African clade; WA: West African clade.
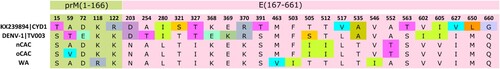
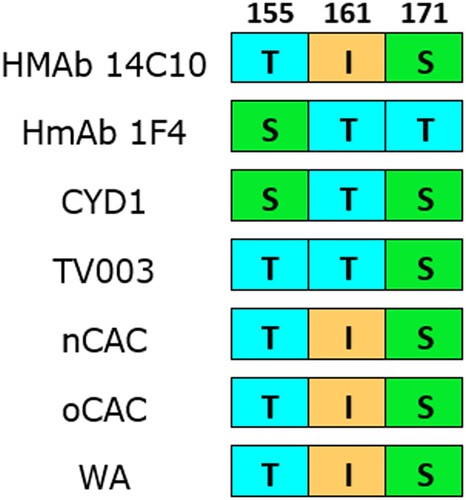
Figure 4. Amino acid divergence comparison between nCAC and epitopes of DENV-1 neutralizing human monoclonal antibodies (HMAb 1F4 and HMAb 14c10) and vaccine strains. Numerals represent the E protein amino acid position. The other African clades were included for comparison. CYD1: Dengvaxia®; TV003: TetraVax-DV-TV003; nCAC: new Central African clade; oCAC: old Central African clade; WA: West African clade.
Data availability
The consensus sequences that were generated from the six Cameroon DENV-1 strains have been deposited at GenBank under accession numbers OQ593388–OQ593393.
