Figures & data
Figure 1. Timeline of the isolation of OXA-232-producing K. pneumoniae during the period of December 2022 to April 2023. A. Timeline of the isolation of OXA-232-producing K. pneumoniae. Coloured rectangles represent the presence of the patient in the corresponding ward. Only patients with isolation of at least two strains were displayed. The yellow and red rectangles represent the detection of ST15 and ST147 OXA-232-producing K. pneumoniae strains, respectively. SICU, surgery intensive care unit; NICU, neurosurgery intensive care unit. B. Linear alignment of blaOXA-232-harbouring ColKP3 plasmids from ST15 and ST147 strains isolated from patient P1. Genes were denoted by arrows, and were coloured based on their functional classification. Shading denotes regions of homology (nucleotide identity ≥99%).
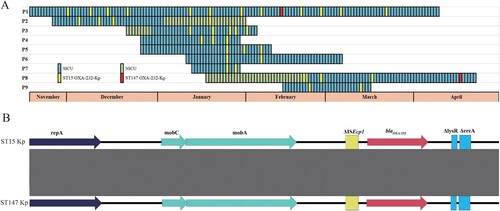
Figure 2. Phylogenetic analysis of OXA-232-producing K. pneumoniae strains. The phylogenetic tree was constructed with core genome sequences of 8 strains collected in this study (ST15 and ST147), and other reference strains retrieved from the GenBank database. The filled block of colour indicates the presence of plasmids and the antimicrobial resistance genes.
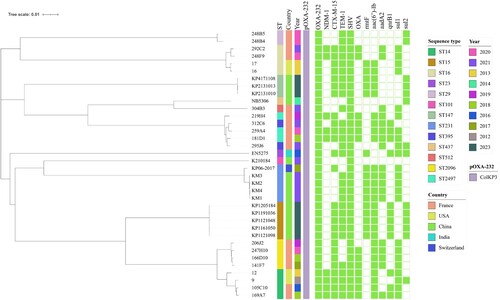
Figure 3. The distribution of plasmids harbouring carbapenemase genes in outer membrane vesicles (OMVs) derived from carbapenemase-producing K. pneumoniae. (A, D, G) Transmission electron microscopy (TEM) imaging of OMVs from OXA-232-Kp, KPC-2-Kp, and NDM-1-Kp. (B, E, H) Nanoparticle tracking analysis (NTA) of OMVs from OXA-232-Kp, KPC-2-Kp, and NDM-1-Kp. (C, F, I) PCR screening for carbapenemase genes and the corresponding plasmid backbones. For each gene, genomic DNA was employed as the positive control and was placed at the left lane. OMVs DNA was also used as the template and the PCR products were run at the right lane. (J) Plasmid copy number of blaOXA-232 ColKP3 (pOXA-232), blaKPC-2 IncFII(pHN7A8)/IncR (pKPC-2), and blaNDM-1 IncX3 (pNDM-1) plasmids in K. pneumoniae. (K) The number of plasmids per nanogram of vesicle protein. (L) The number of plasmids per OMV. The number of OMV was determined with NTA analysis. Error bars signify standard deviations. ****, P < 0.0001.
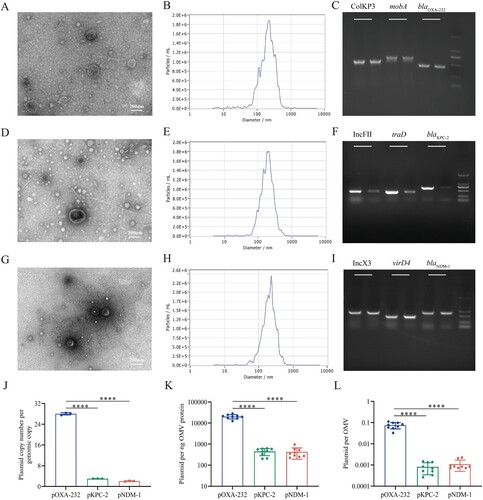
Figure 4. OMVs-mediated transformation assay of blaOXA-232. (A) Protocol of OMVs-mediated transformation assay. (B) Transformants were selected on LB agar plates with 8 µg/ml piperacillin and 4 µg/ml tazobactam. Recipient cells were treated with free plasmid pOXA-232, OMVs and triton-lysed OMVs. Eco, E. coli MG1655 hph ΔhsdR; Kpn, K. pneumoniae TU37-vf Δwza. (C) PCR screening for positive transformants after OMVs mediated transformation assay using E. coli MG1655 hph ΔhsdR and K. pneumoniae TU37-vf Δwza as the recipient strains. (D) Agarose gel electrophoresis of blaOXA-232-harbouring ColKP3 plasmids extracted from E. coli MG1655 hph ΔhsdR (Ec) and K. pneumoniae TU37-vf Δwza (Kp) transformants.
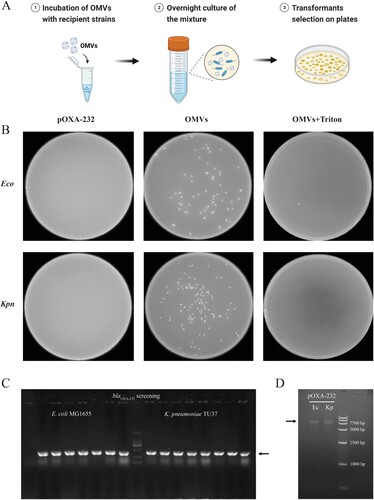
Table 1. Outer membrane vesicles (OMVs) mediated transformation of plasmids-harbouring carbapenemase genes.
Table 2. Antibiotic susceptibility profiles of donor, recipient and OMVs-mediated blaOXA-232 transformants.
