Figures & data
Table 1. MICs of AMXT-1501 against Gram-positive and Gram-negative bacteria.
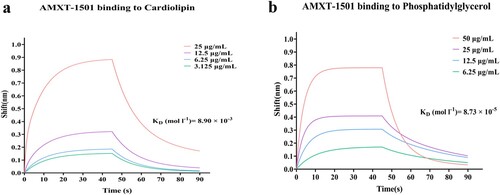
Figure 1. AMXT-1501 has excellent antibacterial activity against MDR S. aureus. (a) Molecular structure and formula of AMXT-1501 (AMXT). (b) Antibacterial activity of AMXT-1501 against MSSA CHS101 and MRSA YUSA145 detected by time-killing assays; compared with vancomycin (VAN); The dashed lines indicated the limit of detection of time-killing assays. Antibacterial activity of AMXT-1501 against skin abscess infected with MRSA YUSA145 (n = 15/group); compared with VAN. (c) The strategy of administration for mice skin abscess model, (d) representative abscesses, (e) lesion areas, and (f) bacteriology in abscesses on day 7. Data in panels b, e, and f were presented as means ± s.d, and P values were determined using an unpaired, two-tailed Student’s t-test. VAN, vancomycin; AMXT, AMXT-1501; MIC, minimum inhibitory concentration; MSSA, methicillin-sensitive S. aureus; MRSA, methicillin-resistant S. aureus;
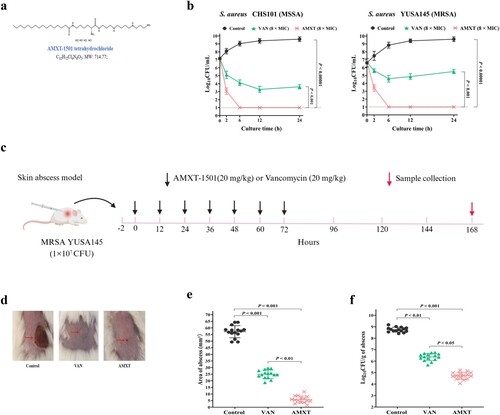
Figure 2. AMXT-1501 has excellent antibacterial activity against Gram-negative MDR bacteria. (a) Antibacterial activity of AMXT-1501 against K. pneumoniae, an ESBL-producing E. coli, and carbapenem-resistant Enterobacteriaceae (CRE) E. coli detected by time-killing assays; compared with meropenem (MER) or tigecycline (TIG); The dashed lines indicated the limit of detection of time-killing assays. Antibacterial activity of AMXT-1501 against abdominal infected with E. coli ECO2219 (n = 15/group); compared with TIG. (b) The strategy of administration for mice abdominal infection model, (c) survival rates, bacteriology in lung (d) and liver (e) on day 7. Data in panels a, d and e were presented as means ± s.d, and P values were determined using an unpaired, two-tailed Student’s t-test. P values were determined using log-rank test for panels c. MER, meropenem; TIG, tigecycline; AMXT, AMXT-1501; MIC, minimum inhibitory concentration; ESBL, extended-spectrum β-lactamase; CRE, carbapenem-resistant Enterobacteriaceae.
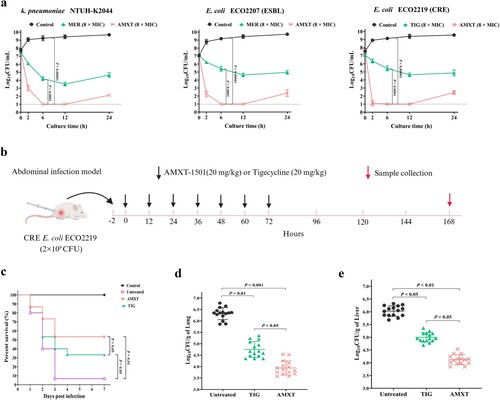
Figure 3. AMXT-1501 reduces the biofilm formation of S. aureus. Effects of sub-MICs of AMXT-1501 on growth of two MSSA (a, c) and two MRSA (b, d) isolates’ planktonic cells determined by optical density at 600 nm (OD600) and as indicated by biofilm biomass determination according to crystal violet staining analysis after 24 h of exposure. Effects of 1/4× MIC of AMXT-1501 on the biofilm formation of eight MSSA (e) and eight MRSA (f) strains determined by crystal violet staining analysis. Graphed data are means ± SDs; *P < 0.05, **P < 0.01. ***P < 0.001 vs. Control (unpaired, two-tailed Student’s t-test). MIC, minimum inhibitory concentration; MSSA, methicillin-sensitive S. aureus; MRSA, methicillin-resistant S. aureus;
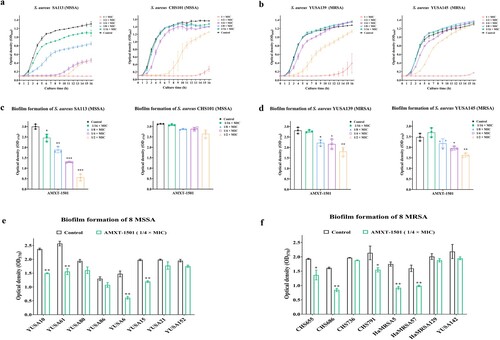
Figure 4. AMXT-1501 destroys bacterial cell membranes of E. coli. (a) E. coli ECO2219 (CRE) was treated with AMXT-1501 (AMXT) for 2 h and observed by SEM. (b) E. coli ECO2219 (CRE) was treated with AMXT-1501 for 2 h, and observed by TEM. Representative fields are shown in all panels. AMXT, AMXT-1501; MIC, minimum inhibitory concentration; CRE, carbapenem-resistant Enterobacteriaceae.
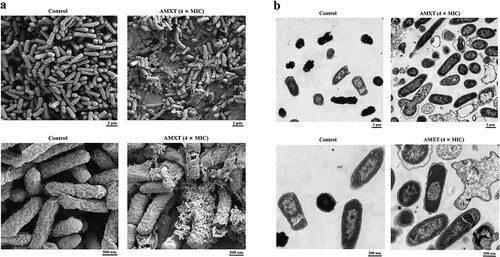
Figure 5. AMXT-1501 increased membrane permeability and depolarization of E. coli. (a) E. coli ECO2219 (CRE) treated with AMXT-1501 for 30 min, labelled with SYTO-9 and PI, and observed under a confocal laser scanning microscope; (b) Fluorescence intensities of PI-labelled E. coli ECO2219 (CRE) treated with AMXT-1501 for 30 min, detected at excitation and emission wavelengths of 535 and 615 nm measured by a microplate reader, respectively. (c) The inner membrane permeabilizing ability of AMXT-1501 was determined by measurement of β-galactosidase activity in E. coli ECO2219 (CRE) using the normally impermeable, chromogenic substrate o-nitrophenyl-β-d-galactoside (ONPG); (d) Fluorescence intensities of DiBAC4(3)-treated E. coli ECO2219 (CRE) treated with AMXT-1501, detected associated fluorescence intensity at 492 and 515 nm for 60 min, respectively. Representative fields are shown in panel a. Data in panels b, c and d were presented as means ± s.d, and P values were determined using an unpaired, two-tailed Student’s t-test. a.u., arbitrary units. AMXT, AMXT-1501; MIC, minimum inhibitory concentration; CRE, carbapenem-resistant Enterobacteriaceae.
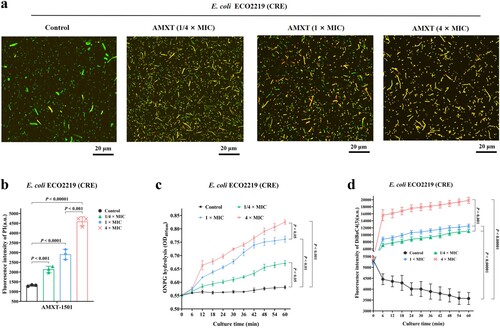
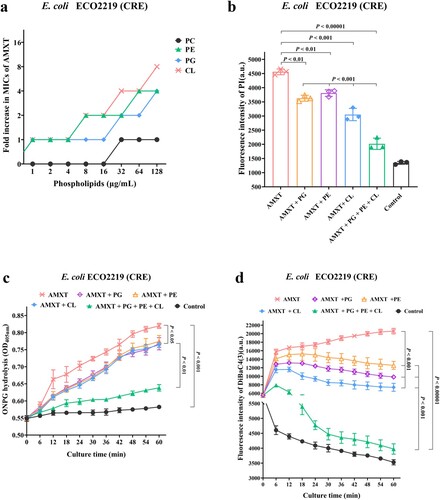
Figure 6. Bactericidal activity of AMXT-1501 via CL and PG against E. coli. (a) Increased MICs of AMXT-1501 against E. coli ECO2219 (CRE) in the presence of PC, PE, PG, and CL; exogenous addition of CL, PG and PE attenuated AMXT-1501 effects on membrane permeability (b, c) and depolarization (d). Data in panels b, c and d were presented as means ± s.d, and P values were determined using an unpaired, two-tailed Student’s t-test. a.u., arbitrary units. AMXT, AMXT-1501; MIC, minimum inhibitory concentration; CRE, carbapenem-resistant Enterobacteriaceae. PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; CL, cardiolipin.
Supplementary_Information
Download PDF (4.3 MB)Data availability statement
The raw whole-genome sequencing data was posted in the Sequence Read Archive (SRA) database under accession number PRJNA1007457 (http://www.ncbi.nlm.nih.gov/bioproject/1007457). All the other data supporting the findings of this study are available within the article and its supplementary information files and from the corresponding authors upon reasonable request. A reporting summary for this article is available as a Supplementary Information file.