Figures & data
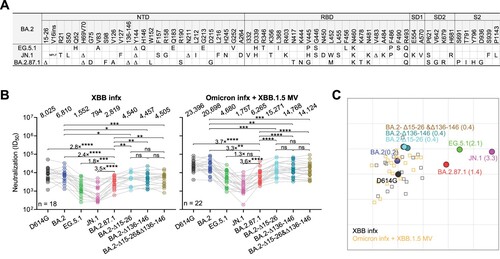
Figure 1. Spike mutations and serum neutralization of SARS-CoV-2 Omicron variant BA.2.87.1. A. Spike mutations of BA.2.87.1 in comparison of EG.5.1 and JN.1 on top of BA.2. Δ, deletion; ins, insertion. B. Neutralizing ID50 titres of serum samples from “XBB infx” and “Omicron infx + XBB.1.5 MV” cohorts against the indicated SARS-CoV-2 variants. The geometric mean ID50 titres (GMT) are presented above symbols. Fold changes in neutralizing ID50 titres between BA.2.87.1 and other SARS-CoV-2 variants are denoted. Statistical analyses were performed by employing Wilcoxon matched-pairs signed-rank tests. ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. n, sample size. Participants from the two cohorts all had received 3–4 doses of the wildtype monovalent vaccines, one dose of the BA.5 bivalent vaccine booster and then followed by either an XBB breakthrough infection (“XBB infx”) or Omicron infection and XBB.1.5 monovalent vaccine booster (“Omicron infx + XBB.1.5 MV”). C. Antigenic map based on the neutralizing data from all serum samples in panel B. D614G represents the central reference for all serum cohorts, with the antigenic distances calculated by the average divergence from each variant. One antigenic unit (AU) represents an approximately 2-fold change in ID50 titre. Variant positions are shown as circles, while sera are denoted as gray and orange squares. Antigenic distances from D614G in antigenic units are in parentheses next to each variant name.